Gene
KWMTBOMO08032
Pre Gene Modal
BGIBMGA001127
Annotation
PREDICTED:_post-GPI_attachment_to_proteins_factor_2-like_[Bombyx_mori]
Full name
Post-GPI attachment to proteins factor 2-like
Location in the cell
PlasmaMembrane Reliability : 4.859
Sequence
CDS
ATGGTAAATGACGCACAATTAAAATCGCCACCAAAATCGGATGTGCACGGAGCGATACGCACAGACAATGATGTAAATATTGATAGGGGTGTTAAAATATTGGAGTACAAGGGCATACTCTGGACATGTGGTTTGCGACAAATATGTTTAGCTGCATTATGGCTTCCGCTGGGTGCTTTAATATTTTGTTACGTGACAGCCTCTATGTTCCAGGCAGATGATATACACGAAACACATTGTAAAGTTTACAATGTGGTCCCGTCAATAAGTGCAATTACCGGTATTAGTCCACAAAGGTATATTTGGAGGATATGCATCGCGTTTCATTTAGGACCAAGATTGTTAATAGGATCCCTGTATTACAATTATCATAAGGAACGAAATAGTTTTATTGGGGATGAAAAGGTACGAGTGCAGGCAAATAAACTGGGAGAGGCTTGCTATTGGCTCAATTTGATAGAGTTATTAGCACTTACGGGCGTCACCTATGTATCTAACAGAGAAAACTATTTTGTGCATGAGAAAATTTTCATAGTTTTTATGATTGTTGCGTTATTGCACATGCTGTGTCGAGGAAGAGTGGGCGCCATCGGAGCTGACTCGGTGACCCCCGTGCGGACCAATAATATAGTCTGGACGCTATTTTACGTGGCTGTGGCGTCGACTGTAGGGCTAGTTGTGTTTTTCTTGAGACACCGCCTTTTGTGCCGACCTCTAGCTTTCACATGGTTCTCAGTCTGCGAATACATTCTGGCTACCACAAACATGGCTTTCCATGCGATTGTGGTGCGCGACCTGCCACTTCACCATCTCCAAGTTGTATATCCTGATCACAAACGAAACTAG
Protein
MVNDAQLKSPPKSDVHGAIRTDNDVNIDRGVKILEYKGILWTCGLRQICLAALWLPLGALIFCYVTASMFQADDIHETHCKVYNVVPSISAITGISPQRYIWRICIAFHLGPRLLIGSLYYNYHKERNSFIGDEKVRVQANKLGEACYWLNLIELLALTGVTYVSNRENYFVHEKIFIVFMIVALLHMLCRGRVGAIGADSVTPVRTNNIVWTLFYVAVASTVGLVVFFLRHRLLCRPLAFTWFSVCEYILATTNMAFHAIVVRDLPLHHLQVVYPDHKRN
Summary
Similarity
Belongs to the PGAP2 family.
Keywords
Complete proteome
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Post-GPI attachment to proteins factor 2-like
Uniprot
A0A2H1WMS1
A0A194Q6H7
A0A194RH70
A0A3S2LT06
A0A212FN71
A0A0L7LK26
+ More
E2BE99 A0A3L8DZN3 E2AY65 A0A0J7L4M2 A0A232EYE4 A0A195FHX0 F4W6M0 A0A195BT91 A0A158P2C0 A0A151WZH1 K7J394 A0A310S624 A0A154P8B7 A0A1B6K390 A0A1B0D232 A0A088AT17 A0A1B6KWS3 V9II09 A0A195E453 A0A1B6D455 A0A1B0C963 A0A067QU53 A0A195D343 A0A1Y1LZG6 Q7QFI7 A0A182Q073 A0A182FJN5 A0A1S4G996 A0A182T1D5 A0A182UDC3 W5JJR9 A0A182VCX6 A0A182Y7T0 E0VDS0 A0A2S2PRS0 A0A2H8TF78 A0A139WMX4 J9K5G6 A0A1J1IAV6 B0XEF9 A0A182J1W4 A0A182W4J5 A0A2P8ZDR7 A0A0L7REW5 A0A2A3EGB2 A0A026WIA9 Q17L81 Q17MC8 A0A0A9XFM9 A0A182MZI3 A0A1A9VRZ8 W8C4G1 A0A1B0GBK3 A0A1A9ZII1 A0A1A9W6E0 A0A1B0BBG9 W8B6X4 A0A1A9XX05 A0A0L0C3K3 A0A0A1WXX5 A0A1W4V2H1 A0A0R3P0T4 B4R7C0 A0A0P5FP14 B4PWT2 B4GVF2 B3NUS9 B4I723 A0A1I8MJM9 Q9VWK6 B5DNA8 B4JKL7 A0A182GZN2 B3N151 A0A1W4X8N8 A0A1W4WZ93 A0A3B0K311 A0A0P6E2U7 A0A0C9PGL1 B4M1T1 A0A182GQU8 B4L8Q9 A0A1I8Q8Q1 B4NBY9 A0A0M3QYZ8 A0A182XE08 A0A034V9Y1 A0A224XIF8 A0A2R5LHF4 A0A0V0G8S7
E2BE99 A0A3L8DZN3 E2AY65 A0A0J7L4M2 A0A232EYE4 A0A195FHX0 F4W6M0 A0A195BT91 A0A158P2C0 A0A151WZH1 K7J394 A0A310S624 A0A154P8B7 A0A1B6K390 A0A1B0D232 A0A088AT17 A0A1B6KWS3 V9II09 A0A195E453 A0A1B6D455 A0A1B0C963 A0A067QU53 A0A195D343 A0A1Y1LZG6 Q7QFI7 A0A182Q073 A0A182FJN5 A0A1S4G996 A0A182T1D5 A0A182UDC3 W5JJR9 A0A182VCX6 A0A182Y7T0 E0VDS0 A0A2S2PRS0 A0A2H8TF78 A0A139WMX4 J9K5G6 A0A1J1IAV6 B0XEF9 A0A182J1W4 A0A182W4J5 A0A2P8ZDR7 A0A0L7REW5 A0A2A3EGB2 A0A026WIA9 Q17L81 Q17MC8 A0A0A9XFM9 A0A182MZI3 A0A1A9VRZ8 W8C4G1 A0A1B0GBK3 A0A1A9ZII1 A0A1A9W6E0 A0A1B0BBG9 W8B6X4 A0A1A9XX05 A0A0L0C3K3 A0A0A1WXX5 A0A1W4V2H1 A0A0R3P0T4 B4R7C0 A0A0P5FP14 B4PWT2 B4GVF2 B3NUS9 B4I723 A0A1I8MJM9 Q9VWK6 B5DNA8 B4JKL7 A0A182GZN2 B3N151 A0A1W4X8N8 A0A1W4WZ93 A0A3B0K311 A0A0P6E2U7 A0A0C9PGL1 B4M1T1 A0A182GQU8 B4L8Q9 A0A1I8Q8Q1 B4NBY9 A0A0M3QYZ8 A0A182XE08 A0A034V9Y1 A0A224XIF8 A0A2R5LHF4 A0A0V0G8S7
Pubmed
26354079
22118469
26227816
20798317
30249741
28648823
+ More
21719571 21347285 20075255 24845553 28004739 12364791 20920257 23761445 25244985 20566863 18362917 19820115 29403074 24508170 17510324 25401762 26823975 24495485 26108605 25830018 15632085 17994087 17550304 25315136 10731132 12537572 12537569 26483478 25348373
21719571 21347285 20075255 24845553 28004739 12364791 20920257 23761445 25244985 20566863 18362917 19820115 29403074 24508170 17510324 25401762 26823975 24495485 26108605 25830018 15632085 17994087 17550304 25315136 10731132 12537572 12537569 26483478 25348373
EMBL
ODYU01009740
SOQ54370.1
KQ459439
KPJ00979.1
KQ460397
KPJ15281.1
+ More
RSAL01000232 RVE43817.1 AGBW02006772 OWR55192.1 JTDY01000816 KOB75775.1 GL447768 EFN85944.1 QOIP01000002 RLU25762.1 GL443762 EFN61661.1 LBMM01000757 KMQ97561.1 NNAY01001629 OXU23381.1 KQ981523 KYN40290.1 GL887707 EGI70355.1 KQ976408 KYM91156.1 ADTU01007083 KQ982639 KYQ53269.1 KQ768935 OAD53027.1 KQ434841 KZC08091.1 GECU01019653 GECU01019153 GECU01002063 JAS88053.1 JAS88553.1 JAT05644.1 AJVK01002639 AJVK01002640 GEBQ01024078 JAT15899.1 JR048487 AEY60680.1 KQ979657 KYN19965.1 GEDC01016826 JAS20472.1 AJWK01001948 KK853265 KDR09196.1 KQ976885 KYN07317.1 GEZM01044165 JAV78481.1 AAAB01008846 EAA06607.2 AXCN02001274 ADMH02001027 ETN64371.1 DS235083 EEB11526.1 GGMR01019448 MBY32067.1 GFXV01000929 MBW12734.1 KQ971312 KYB29368.1 ABLF02036875 ABLF02036878 CVRI01000044 CRK96866.1 DS232826 DS233497 EDS25956.1 EDS30266.1 PYGN01000086 PSN54652.1 KQ414609 KOC69281.1 KZ288254 PBC30750.1 KK107207 EZA55401.1 CH477217 EAT47494.1 CH477207 EAT47821.1 GBHO01025138 GDHC01012439 JAG18466.1 JAQ06190.1 GAMC01009621 JAB96934.1 CCAG010001810 JXJN01011439 GAMC01009620 JAB96935.1 JRES01000948 KNC26885.1 GBXI01010776 JAD03516.1 CH379064 KRT06833.1 CM000366 EDX18361.1 GDIQ01254189 GDIP01135043 LRGB01003146 JAJ97535.1 JAL68671.1 KZS04038.1 CM000162 EDX02816.2 CH479192 EDW26439.1 CH954180 EDV46676.1 CH480823 EDW56121.1 AE014298 BT031012 BT031025 EDY72915.1 CH916370 EDW00120.1 JXUM01099925 JXUM01099926 KQ564528 KXJ72134.1 CH902650 EDV30086.1 OUUW01000015 SPP88647.1 GDIQ01069423 JAN25314.1 GBYB01000006 GBYB01000008 JAG69773.1 JAG69775.1 CH940651 EDW65635.2 JXUM01081370 JXUM01081371 KQ563242 KXJ74223.1 CH933815 EDW08034.2 CH964239 EDW82348.1 CP012528 ALC48553.1 GAKP01020594 JAC38358.1 GFTR01004161 JAW12265.1 GGLE01004816 MBY08942.1 GECL01002004 JAP04120.1
RSAL01000232 RVE43817.1 AGBW02006772 OWR55192.1 JTDY01000816 KOB75775.1 GL447768 EFN85944.1 QOIP01000002 RLU25762.1 GL443762 EFN61661.1 LBMM01000757 KMQ97561.1 NNAY01001629 OXU23381.1 KQ981523 KYN40290.1 GL887707 EGI70355.1 KQ976408 KYM91156.1 ADTU01007083 KQ982639 KYQ53269.1 KQ768935 OAD53027.1 KQ434841 KZC08091.1 GECU01019653 GECU01019153 GECU01002063 JAS88053.1 JAS88553.1 JAT05644.1 AJVK01002639 AJVK01002640 GEBQ01024078 JAT15899.1 JR048487 AEY60680.1 KQ979657 KYN19965.1 GEDC01016826 JAS20472.1 AJWK01001948 KK853265 KDR09196.1 KQ976885 KYN07317.1 GEZM01044165 JAV78481.1 AAAB01008846 EAA06607.2 AXCN02001274 ADMH02001027 ETN64371.1 DS235083 EEB11526.1 GGMR01019448 MBY32067.1 GFXV01000929 MBW12734.1 KQ971312 KYB29368.1 ABLF02036875 ABLF02036878 CVRI01000044 CRK96866.1 DS232826 DS233497 EDS25956.1 EDS30266.1 PYGN01000086 PSN54652.1 KQ414609 KOC69281.1 KZ288254 PBC30750.1 KK107207 EZA55401.1 CH477217 EAT47494.1 CH477207 EAT47821.1 GBHO01025138 GDHC01012439 JAG18466.1 JAQ06190.1 GAMC01009621 JAB96934.1 CCAG010001810 JXJN01011439 GAMC01009620 JAB96935.1 JRES01000948 KNC26885.1 GBXI01010776 JAD03516.1 CH379064 KRT06833.1 CM000366 EDX18361.1 GDIQ01254189 GDIP01135043 LRGB01003146 JAJ97535.1 JAL68671.1 KZS04038.1 CM000162 EDX02816.2 CH479192 EDW26439.1 CH954180 EDV46676.1 CH480823 EDW56121.1 AE014298 BT031012 BT031025 EDY72915.1 CH916370 EDW00120.1 JXUM01099925 JXUM01099926 KQ564528 KXJ72134.1 CH902650 EDV30086.1 OUUW01000015 SPP88647.1 GDIQ01069423 JAN25314.1 GBYB01000006 GBYB01000008 JAG69773.1 JAG69775.1 CH940651 EDW65635.2 JXUM01081370 JXUM01081371 KQ563242 KXJ74223.1 CH933815 EDW08034.2 CH964239 EDW82348.1 CP012528 ALC48553.1 GAKP01020594 JAC38358.1 GFTR01004161 JAW12265.1 GGLE01004816 MBY08942.1 GECL01002004 JAP04120.1
Proteomes
UP000053268
UP000053240
UP000283053
UP000007151
UP000037510
UP000008237
+ More
UP000279307 UP000000311 UP000036403 UP000215335 UP000078541 UP000007755 UP000078540 UP000005205 UP000075809 UP000002358 UP000076502 UP000092462 UP000005203 UP000078492 UP000092461 UP000027135 UP000078542 UP000007062 UP000075886 UP000069272 UP000075901 UP000075902 UP000000673 UP000075903 UP000076408 UP000009046 UP000007266 UP000007819 UP000183832 UP000002320 UP000075880 UP000075920 UP000245037 UP000053825 UP000242457 UP000053097 UP000008820 UP000075884 UP000078200 UP000092444 UP000092445 UP000091820 UP000092460 UP000092443 UP000037069 UP000192221 UP000001819 UP000000304 UP000076858 UP000002282 UP000008744 UP000008711 UP000001292 UP000095301 UP000000803 UP000001070 UP000069940 UP000249989 UP000007801 UP000192223 UP000268350 UP000008792 UP000009192 UP000095300 UP000007798 UP000092553 UP000076407
UP000279307 UP000000311 UP000036403 UP000215335 UP000078541 UP000007755 UP000078540 UP000005205 UP000075809 UP000002358 UP000076502 UP000092462 UP000005203 UP000078492 UP000092461 UP000027135 UP000078542 UP000007062 UP000075886 UP000069272 UP000075901 UP000075902 UP000000673 UP000075903 UP000076408 UP000009046 UP000007266 UP000007819 UP000183832 UP000002320 UP000075880 UP000075920 UP000245037 UP000053825 UP000242457 UP000053097 UP000008820 UP000075884 UP000078200 UP000092444 UP000092445 UP000091820 UP000092460 UP000092443 UP000037069 UP000192221 UP000001819 UP000000304 UP000076858 UP000002282 UP000008744 UP000008711 UP000001292 UP000095301 UP000000803 UP000001070 UP000069940 UP000249989 UP000007801 UP000192223 UP000268350 UP000008792 UP000009192 UP000095300 UP000007798 UP000092553 UP000076407
PRIDE
Pfam
PF10277 Frag1
ProteinModelPortal
A0A2H1WMS1
A0A194Q6H7
A0A194RH70
A0A3S2LT06
A0A212FN71
A0A0L7LK26
+ More
E2BE99 A0A3L8DZN3 E2AY65 A0A0J7L4M2 A0A232EYE4 A0A195FHX0 F4W6M0 A0A195BT91 A0A158P2C0 A0A151WZH1 K7J394 A0A310S624 A0A154P8B7 A0A1B6K390 A0A1B0D232 A0A088AT17 A0A1B6KWS3 V9II09 A0A195E453 A0A1B6D455 A0A1B0C963 A0A067QU53 A0A195D343 A0A1Y1LZG6 Q7QFI7 A0A182Q073 A0A182FJN5 A0A1S4G996 A0A182T1D5 A0A182UDC3 W5JJR9 A0A182VCX6 A0A182Y7T0 E0VDS0 A0A2S2PRS0 A0A2H8TF78 A0A139WMX4 J9K5G6 A0A1J1IAV6 B0XEF9 A0A182J1W4 A0A182W4J5 A0A2P8ZDR7 A0A0L7REW5 A0A2A3EGB2 A0A026WIA9 Q17L81 Q17MC8 A0A0A9XFM9 A0A182MZI3 A0A1A9VRZ8 W8C4G1 A0A1B0GBK3 A0A1A9ZII1 A0A1A9W6E0 A0A1B0BBG9 W8B6X4 A0A1A9XX05 A0A0L0C3K3 A0A0A1WXX5 A0A1W4V2H1 A0A0R3P0T4 B4R7C0 A0A0P5FP14 B4PWT2 B4GVF2 B3NUS9 B4I723 A0A1I8MJM9 Q9VWK6 B5DNA8 B4JKL7 A0A182GZN2 B3N151 A0A1W4X8N8 A0A1W4WZ93 A0A3B0K311 A0A0P6E2U7 A0A0C9PGL1 B4M1T1 A0A182GQU8 B4L8Q9 A0A1I8Q8Q1 B4NBY9 A0A0M3QYZ8 A0A182XE08 A0A034V9Y1 A0A224XIF8 A0A2R5LHF4 A0A0V0G8S7
E2BE99 A0A3L8DZN3 E2AY65 A0A0J7L4M2 A0A232EYE4 A0A195FHX0 F4W6M0 A0A195BT91 A0A158P2C0 A0A151WZH1 K7J394 A0A310S624 A0A154P8B7 A0A1B6K390 A0A1B0D232 A0A088AT17 A0A1B6KWS3 V9II09 A0A195E453 A0A1B6D455 A0A1B0C963 A0A067QU53 A0A195D343 A0A1Y1LZG6 Q7QFI7 A0A182Q073 A0A182FJN5 A0A1S4G996 A0A182T1D5 A0A182UDC3 W5JJR9 A0A182VCX6 A0A182Y7T0 E0VDS0 A0A2S2PRS0 A0A2H8TF78 A0A139WMX4 J9K5G6 A0A1J1IAV6 B0XEF9 A0A182J1W4 A0A182W4J5 A0A2P8ZDR7 A0A0L7REW5 A0A2A3EGB2 A0A026WIA9 Q17L81 Q17MC8 A0A0A9XFM9 A0A182MZI3 A0A1A9VRZ8 W8C4G1 A0A1B0GBK3 A0A1A9ZII1 A0A1A9W6E0 A0A1B0BBG9 W8B6X4 A0A1A9XX05 A0A0L0C3K3 A0A0A1WXX5 A0A1W4V2H1 A0A0R3P0T4 B4R7C0 A0A0P5FP14 B4PWT2 B4GVF2 B3NUS9 B4I723 A0A1I8MJM9 Q9VWK6 B5DNA8 B4JKL7 A0A182GZN2 B3N151 A0A1W4X8N8 A0A1W4WZ93 A0A3B0K311 A0A0P6E2U7 A0A0C9PGL1 B4M1T1 A0A182GQU8 B4L8Q9 A0A1I8Q8Q1 B4NBY9 A0A0M3QYZ8 A0A182XE08 A0A034V9Y1 A0A224XIF8 A0A2R5LHF4 A0A0V0G8S7
Ontologies
PANTHER
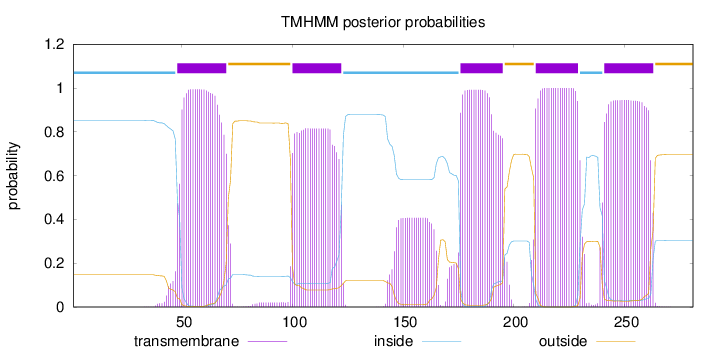
Topology
Subcellular location
Membrane
Length:
281
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
112.08904
Exp number, first 60 AAs:
12.43005
Total prob of N-in:
0.85246
POSSIBLE N-term signal
sequence
inside
1 - 47
TMhelix
48 - 70
outside
71 - 99
TMhelix
100 - 122
inside
123 - 175
TMhelix
176 - 195
outside
196 - 209
TMhelix
210 - 229
inside
230 - 240
TMhelix
241 - 263
outside
264 - 281
Population Genetic Test Statistics
Pi
18.25844
Theta
17.861101
Tajima's D
0.042557
CLR
0
CSRT
0.387630618469077
Interpretation
Uncertain
