Gene
KWMTBOMO08030
Pre Gene Modal
BGIBMGA001129
Annotation
PREDICTED:_glucosylceramidase-like_[Amyelois_transitella]
Full name
Glucosylceramidase
Location in the cell
Extracellular Reliability : 1.047 Nuclear Reliability : 1.282
Sequence
CDS
ATGACGGACTTGAACAACTTCGTCGTCCGCTGGATAGACTGGAATCTGTGTCTCGACCCTGAAGGTGGTCCAAACTGGGCCGATAATTTCGTTGACTCTCCCATCTTAGTGTACGGTGACAAAGATCAATTTATCAAGCAGCCAATGTTCTATGCCATGGGACACTTCTCCAAGTTCATATACAGAGGCTCAAGACGGATTCAGGTATCGCGTAGGAGTCTCGCTCCAATAGAGAACGTGGGACTTTTGACGCCCAACGAGGAGAACCTTTTGTTGGTCTTACAAAACAGGAGACAAAGAGAAGGCTAA
Protein
MTDLNNFVVRWIDWNLCLDPEGGPNWADNFVDSPILVYGDKDQFIKQPMFYAMGHFSKFIYRGSRRIQVSRRSLAPIENVGLLTPNEENLLLVLQNRRQREG
Summary
Catalytic Activity
a beta-D-glucosyl-(1<->1')-N-acylsphing-4-enine + H2O = an N-acylsphing-4-enine + D-glucose
Similarity
Belongs to the glycosyl hydrolase 30 family.
Feature
chain Glucosylceramidase
Uniprot
H9IV49
A0A212F349
A0A0L7L523
A0A194RBR5
A0A194Q614
A0A2H1VA54
+ More
A0A3S2LNQ5 A0A0N1INL2 A0A1W1EGM7 H9JUB6 A0A212EPK9 A0A212F248 A0A2A4K8P8 A0A091FL64 A0A194PPY1 A0A091IT00 A0A3M6UIQ7 A0A087QSU3 A0A0N1IF15 A0A0Q3RD78 J9L5B0 A0A146N104 A0A0M3JU31 A0A2I0TLG3 A0A091QVX9 A0A2H1VG50 A0A3M0IZB8 A0A0N4UDD2 A0A2S2P4H2 A0A093B9F4 A0A147A6F7 A0A2D4JB19 A0A0B1THP5 H9JUB7 A0A2D0R7B2 W5ULT4 A0A3Q2Q153 G3W179 G3PDK2 G3PDK9 A0A146QYI1 A0A3Q2PZJ7 A0A2G9U5T9 I3JGM4 A0A0B1RX03 A0A2I4CSF9 A0A3B4XWH4 A0A218UMY8 A0A146PRV1 A0A3B3ZY58 A0A368H9P3 A0A2H1VUS0 A0A3B3ZYK5 A0A0M3HW45 A0A0B1TDP7 A0A2K6AH77 A0A0Q3QSH3 A0A1A8J899 A0A093G412 A0A1I7XHV7 A0A1A8CNT0 A0A1A8EGL1 F6VKK2 A0A1A8HCI6 A0A1A8EHM4 A0A3B1J6P4 A0A1A8VEJ5 A0A0B2VJ74 A0A2K6AH31 H0XZ17 A0A2K6B2N5 A0A0S7G1W2 A0A2K6AH39 A0A212F254 A0A3Q4G8X5 A0A250YJK2 A0A3S2PC18 A0A3B1IXM1 A0A3P9I394 A0A2K6AH87 A0A2K6B2V9 A0A0F8AEA4 A0A3Q4G8W1 A0A2K6AH60
A0A3S2LNQ5 A0A0N1INL2 A0A1W1EGM7 H9JUB6 A0A212EPK9 A0A212F248 A0A2A4K8P8 A0A091FL64 A0A194PPY1 A0A091IT00 A0A3M6UIQ7 A0A087QSU3 A0A0N1IF15 A0A0Q3RD78 J9L5B0 A0A146N104 A0A0M3JU31 A0A2I0TLG3 A0A091QVX9 A0A2H1VG50 A0A3M0IZB8 A0A0N4UDD2 A0A2S2P4H2 A0A093B9F4 A0A147A6F7 A0A2D4JB19 A0A0B1THP5 H9JUB7 A0A2D0R7B2 W5ULT4 A0A3Q2Q153 G3W179 G3PDK2 G3PDK9 A0A146QYI1 A0A3Q2PZJ7 A0A2G9U5T9 I3JGM4 A0A0B1RX03 A0A2I4CSF9 A0A3B4XWH4 A0A218UMY8 A0A146PRV1 A0A3B3ZY58 A0A368H9P3 A0A2H1VUS0 A0A3B3ZYK5 A0A0M3HW45 A0A0B1TDP7 A0A2K6AH77 A0A0Q3QSH3 A0A1A8J899 A0A093G412 A0A1I7XHV7 A0A1A8CNT0 A0A1A8EGL1 F6VKK2 A0A1A8HCI6 A0A1A8EHM4 A0A3B1J6P4 A0A1A8VEJ5 A0A0B2VJ74 A0A2K6AH31 H0XZ17 A0A2K6B2N5 A0A0S7G1W2 A0A2K6AH39 A0A212F254 A0A3Q4G8X5 A0A250YJK2 A0A3S2PC18 A0A3B1IXM1 A0A3P9I394 A0A2K6AH87 A0A2K6B2V9 A0A0F8AEA4 A0A3Q4G8W1 A0A2K6AH60
EC Number
3.2.1.45
Pubmed
EMBL
BABH01000464
BABH01000465
AGBW02010629
OWR48154.1
JTDY01002868
KOB70572.1
+ More
KQ460397 KPJ15278.1 KQ459439 KPJ00977.1 ODYU01001475 SOQ37718.1 RSAL01002744 RVE40426.1 KQ461178 KPJ07772.1 LT635665 SGZ49395.1 BABH01038539 BABH01038540 BABH01038541 AGBW02013448 OWR43423.1 AGBW02010783 OWR47794.1 NWSH01000035 PCG80449.1 KL447118 KFO69884.1 KQ459602 KPI93190.1 KL218155 KFP02781.1 RCHS01001420 RMX53570.1 KL225872 KFM04297.1 KPJ07773.1 LMAW01001474 KQK83441.1 ABLF02008893 GCES01160813 JAQ25509.1 UYRR01031043 VDK44415.1 KZ508914 PKU34658.1 KK683358 KFQ13519.1 ODYU01002190 SOQ39362.1 QRBI01000202 RMB93898.1 UYYG01001177 VDN59158.1 GGMR01011734 MBY24353.1 KN125716 KFU83959.1 GCES01012212 JAR74111.1 IACK01152512 LAA93584.1 KN549912 KHJ95601.1 BABH01038531 BABH01038532 BABH01038533 BABH01038534 JT411533 JT418303 AHH42685.1 AEFK01161925 GCES01125017 JAQ61305.1 KZ349327 PIO65072.1 AERX01025232 KN610588 KHJ77613.1 MUZQ01000219 OWK55104.1 GCES01139623 JAQ46699.1 JOJR01000002 RCN53333.1 ODYU01004571 SOQ44585.1 KN550413 KHJ94226.1 LMAW01002883 KQK76089.1 HAED01019445 HAEE01008693 SBR05907.1 KL215229 KFV63808.1 HADZ01017496 SBP81437.1 HAEA01016548 SBQ45029.1 HAEC01013684 SBQ81901.1 HAEB01000173 SBQ46571.1 HADY01022016 HAEJ01018383 SBS58840.1 JPKZ01001512 UYWY01023004 KHN81497.1 VDM46971.1 AAQR03119144 AAQR03119145 GBYX01458341 JAO23212.1 OWR47828.1 GFFW01001008 JAV43780.1 CM012452 RVE62840.1 KQ042766 KKF10976.1
KQ460397 KPJ15278.1 KQ459439 KPJ00977.1 ODYU01001475 SOQ37718.1 RSAL01002744 RVE40426.1 KQ461178 KPJ07772.1 LT635665 SGZ49395.1 BABH01038539 BABH01038540 BABH01038541 AGBW02013448 OWR43423.1 AGBW02010783 OWR47794.1 NWSH01000035 PCG80449.1 KL447118 KFO69884.1 KQ459602 KPI93190.1 KL218155 KFP02781.1 RCHS01001420 RMX53570.1 KL225872 KFM04297.1 KPJ07773.1 LMAW01001474 KQK83441.1 ABLF02008893 GCES01160813 JAQ25509.1 UYRR01031043 VDK44415.1 KZ508914 PKU34658.1 KK683358 KFQ13519.1 ODYU01002190 SOQ39362.1 QRBI01000202 RMB93898.1 UYYG01001177 VDN59158.1 GGMR01011734 MBY24353.1 KN125716 KFU83959.1 GCES01012212 JAR74111.1 IACK01152512 LAA93584.1 KN549912 KHJ95601.1 BABH01038531 BABH01038532 BABH01038533 BABH01038534 JT411533 JT418303 AHH42685.1 AEFK01161925 GCES01125017 JAQ61305.1 KZ349327 PIO65072.1 AERX01025232 KN610588 KHJ77613.1 MUZQ01000219 OWK55104.1 GCES01139623 JAQ46699.1 JOJR01000002 RCN53333.1 ODYU01004571 SOQ44585.1 KN550413 KHJ94226.1 LMAW01002883 KQK76089.1 HAED01019445 HAEE01008693 SBR05907.1 KL215229 KFV63808.1 HADZ01017496 SBP81437.1 HAEA01016548 SBQ45029.1 HAEC01013684 SBQ81901.1 HAEB01000173 SBQ46571.1 HADY01022016 HAEJ01018383 SBS58840.1 JPKZ01001512 UYWY01023004 KHN81497.1 VDM46971.1 AAQR03119144 AAQR03119145 GBYX01458341 JAO23212.1 OWR47828.1 GFFW01001008 JAV43780.1 CM012452 RVE62840.1 KQ042766 KKF10976.1
Proteomes
UP000005204
UP000007151
UP000037510
UP000053240
UP000053268
UP000283053
+ More
UP000218220 UP000053760 UP000054308 UP000275408 UP000053286 UP000051836 UP000007819 UP000036680 UP000269221 UP000038040 UP000274756 UP000221080 UP000265000 UP000007648 UP000007635 UP000005207 UP000192220 UP000261360 UP000197619 UP000261520 UP000252519 UP000036681 UP000233140 UP000053875 UP000095283 UP000002280 UP000018467 UP000031036 UP000050794 UP000267007 UP000005225 UP000233120 UP000261580 UP000265200
UP000218220 UP000053760 UP000054308 UP000275408 UP000053286 UP000051836 UP000007819 UP000036680 UP000269221 UP000038040 UP000274756 UP000221080 UP000265000 UP000007648 UP000007635 UP000005207 UP000192220 UP000261360 UP000197619 UP000261520 UP000252519 UP000036681 UP000233140 UP000053875 UP000095283 UP000002280 UP000018467 UP000031036 UP000050794 UP000267007 UP000005225 UP000233120 UP000261580 UP000265200
Pfam
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
H9IV49
A0A212F349
A0A0L7L523
A0A194RBR5
A0A194Q614
A0A2H1VA54
+ More
A0A3S2LNQ5 A0A0N1INL2 A0A1W1EGM7 H9JUB6 A0A212EPK9 A0A212F248 A0A2A4K8P8 A0A091FL64 A0A194PPY1 A0A091IT00 A0A3M6UIQ7 A0A087QSU3 A0A0N1IF15 A0A0Q3RD78 J9L5B0 A0A146N104 A0A0M3JU31 A0A2I0TLG3 A0A091QVX9 A0A2H1VG50 A0A3M0IZB8 A0A0N4UDD2 A0A2S2P4H2 A0A093B9F4 A0A147A6F7 A0A2D4JB19 A0A0B1THP5 H9JUB7 A0A2D0R7B2 W5ULT4 A0A3Q2Q153 G3W179 G3PDK2 G3PDK9 A0A146QYI1 A0A3Q2PZJ7 A0A2G9U5T9 I3JGM4 A0A0B1RX03 A0A2I4CSF9 A0A3B4XWH4 A0A218UMY8 A0A146PRV1 A0A3B3ZY58 A0A368H9P3 A0A2H1VUS0 A0A3B3ZYK5 A0A0M3HW45 A0A0B1TDP7 A0A2K6AH77 A0A0Q3QSH3 A0A1A8J899 A0A093G412 A0A1I7XHV7 A0A1A8CNT0 A0A1A8EGL1 F6VKK2 A0A1A8HCI6 A0A1A8EHM4 A0A3B1J6P4 A0A1A8VEJ5 A0A0B2VJ74 A0A2K6AH31 H0XZ17 A0A2K6B2N5 A0A0S7G1W2 A0A2K6AH39 A0A212F254 A0A3Q4G8X5 A0A250YJK2 A0A3S2PC18 A0A3B1IXM1 A0A3P9I394 A0A2K6AH87 A0A2K6B2V9 A0A0F8AEA4 A0A3Q4G8W1 A0A2K6AH60
A0A3S2LNQ5 A0A0N1INL2 A0A1W1EGM7 H9JUB6 A0A212EPK9 A0A212F248 A0A2A4K8P8 A0A091FL64 A0A194PPY1 A0A091IT00 A0A3M6UIQ7 A0A087QSU3 A0A0N1IF15 A0A0Q3RD78 J9L5B0 A0A146N104 A0A0M3JU31 A0A2I0TLG3 A0A091QVX9 A0A2H1VG50 A0A3M0IZB8 A0A0N4UDD2 A0A2S2P4H2 A0A093B9F4 A0A147A6F7 A0A2D4JB19 A0A0B1THP5 H9JUB7 A0A2D0R7B2 W5ULT4 A0A3Q2Q153 G3W179 G3PDK2 G3PDK9 A0A146QYI1 A0A3Q2PZJ7 A0A2G9U5T9 I3JGM4 A0A0B1RX03 A0A2I4CSF9 A0A3B4XWH4 A0A218UMY8 A0A146PRV1 A0A3B3ZY58 A0A368H9P3 A0A2H1VUS0 A0A3B3ZYK5 A0A0M3HW45 A0A0B1TDP7 A0A2K6AH77 A0A0Q3QSH3 A0A1A8J899 A0A093G412 A0A1I7XHV7 A0A1A8CNT0 A0A1A8EGL1 F6VKK2 A0A1A8HCI6 A0A1A8EHM4 A0A3B1J6P4 A0A1A8VEJ5 A0A0B2VJ74 A0A2K6AH31 H0XZ17 A0A2K6B2N5 A0A0S7G1W2 A0A2K6AH39 A0A212F254 A0A3Q4G8X5 A0A250YJK2 A0A3S2PC18 A0A3B1IXM1 A0A3P9I394 A0A2K6AH87 A0A2K6B2V9 A0A0F8AEA4 A0A3Q4G8W1 A0A2K6AH60
PDB
2XWE
E-value=5.21078e-22,
Score=251
Ontologies
GO
GO:0003700
GO:0004348
GO:0006665
GO:0016021
GO:0036268
GO:0030278
GO:0070050
GO:0005623
GO:0043243
GO:0005765
GO:0043202
GO:1905037
GO:1903061
GO:0006680
GO:0009267
GO:0046512
GO:0032463
GO:0007040
GO:0005783
GO:0005615
GO:0032436
GO:0032715
GO:0050295
GO:1904457
GO:0005802
GO:0005124
GO:0046527
GO:0030259
GO:0071356
GO:0023021
GO:0035307
GO:0046513
GO:0008203
GO:1901215
GO:0043407
GO:0032006
GO:1904925
GO:0016787
GO:0005975
GO:0016020
GO:0005524
GO:0006412
GO:0016876
GO:0043039
PANTHER
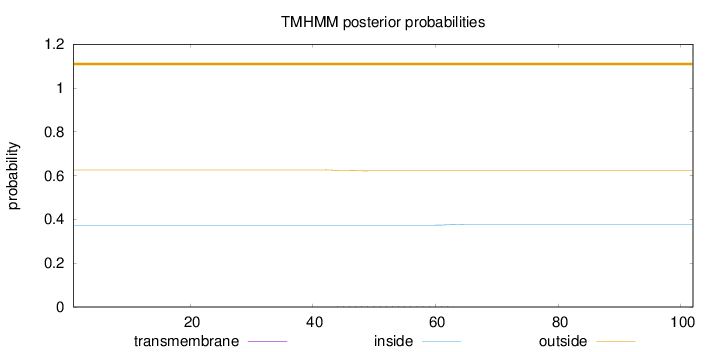
Topology
Length:
102
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0808200000000001
Exp number, first 60 AAs:
0.07005
Total prob of N-in:
0.37384
outside
1 - 102
Population Genetic Test Statistics
Pi
33.262151
Theta
31.669372
Tajima's D
0.294719
CLR
0.573367
CSRT
0.461276936153192
Interpretation
Uncertain
