Gene
KWMTBOMO08029
Pre Gene Modal
BGIBMGA001129
Annotation
PREDICTED:_glucosylceramidase-like_[Bombyx_mori]
Full name
Glucosylceramidase
Location in the cell
Cytoplasmic Reliability : 1.815
Sequence
CDS
ATGGCATACACTAGTACCAATGACGGACTTCGTTTCTCTAAAAGTCAGGGTCAAGTTACGCACGATGAACAATTTGATGCTTCGGTCGCTGAAAAACACGAGAGTTTGCCTAAAGAATTCGATTTCTTCACGACTGGTGTTGTTTTAAGAGTGGACAGCACCATCAAGTTTCAAACAATAGAAGGATTCGGTGGAGCGGTAACCGATGCTGCGTCTTTGAACTGGAGGAAGCTTCCACCGGCCGCACAGGATAAGCTAATCGAAACCTACTTCGGTCCAAATGGCTTGGAATATAATATGATGCGAGTACCAATCGGCGGATCTGACTTTTCAACCCACCCTTACACTTACAATGAACAACCCTGGAACGATACTGAACTCTCGAATTTCTCCCTAACGAATGAGGATATTTCTTTCAAGATACCAATGATTCAGAATTGTATAAAGGCAGCGACTAGTGAGATACACATAACCGCTACAACGTGGTCGCCTCCGGTTTGGATGAAAACAAACGAAAAAATATCTGGCTATGGACAGCTCAAAACCGAATATTACCAGATTTACGCAGATTATCATTTGCGATTCTTGGAAGAATATAAGAAACACGACATCCATATATGGGCCATCACTACAACTAATGAACCAGTCAATGGTATCATACCGGTGGTTACGTTCAATTCTATGGGATGGATTCCTTCGCAATTAGGTCGTTGGATAGAAAACAACCTCGGCCCTACAATAAGGAAGTCTGTTTTCAATAGAACATTAATTCTTGGTGTTGACGACCAGAGGCTGATGCTGTCTGCTTACATGCTAGGAATGGAGAAAGCCGCACCAAACTCCATCAATTATTTAGATGGTATAGCAGTACATTATTATGAAGATTTCGTTTCACCTGAAATTCTTACAAATCTACACAAACGTTATCCAACGAAAATTATAGTTGCCACAGAAGCATGTGAAGGTGCGTATCCTTGGGAAACCTCTAAAGTTAAGATCGGATCTTGGGACAGAGCACAGAACTATGTCAAAGACATTATAGAGGTAATTCATAAATTGTTGTAA
Protein
MAYTSTNDGLRFSKSQGQVTHDEQFDASVAEKHESLPKEFDFFTTGVVLRVDSTIKFQTIEGFGGAVTDAASLNWRKLPPAAQDKLIETYFGPNGLEYNMMRVPIGGSDFSTHPYTYNEQPWNDTELSNFSLTNEDISFKIPMIQNCIKAATSEIHITATTWSPPVWMKTNEKISGYGQLKTEYYQIYADYHLRFLEEYKKHDIHIWAITTTNEPVNGIIPVVTFNSMGWIPSQLGRWIENNLGPTIRKSVFNRTLILGVDDQRLMLSAYMLGMEKAAPNSINYLDGIAVHYYEDFVSPEILTNLHKRYPTKIIVATEACEGAYPWETSKVKIGSWDRAQNYVKDIIEVIHKLL
Summary
Catalytic Activity
a beta-D-glucosyl-(1<->1')-N-acylsphing-4-enine + H2O = an N-acylsphing-4-enine + D-glucose
Similarity
Belongs to the glycosyl hydrolase 30 family.
Feature
chain Glucosylceramidase
Uniprot
A0A194Q614
A0A212FNA8
A0A194PPY1
H9JUB6
A0A212F248
A0A194RBR5
+ More
A0A2H1VUS0 A0A1W1EGM7 A0A1B6LUZ4 A0A1B6IGC1 A0A1B6K1N7 A0A3S2NLJ3 A0A1B6FTD6 A0A2A4K975 J9K5F8 A0A2J7R6I8 A0A1B6J3H7 A0A2J7R6J3 A0A2H1VST3 A0A2H1X380 A0A2J7R6J1 A0A2H1VG50 A0A0P5UEK4 A0A2A4K8P8 A0A212F254 A0A0P5UKE0 A0A0N8CP26 A0A0N1IF15 A0A0P5V259 A0A0P5V115 A0A067QM95 A0A0P5VAG5 A0A154P4F4 A0A0P5D4A0 A0A0N8BXP2 A0A0N8C3M9 A0A0P6HWY3 A0A2A3EMA9 A0A0L7QMS7 A0A0P6CTA3 A0A212F293 A0A0C9QXG4 A0A0P5BCG5 H9JUB7 A0A0P5UX07 A0A0P5YE76 A0A088APM4 A0A0P5BE18 T1JDD3 I4DLH0 A0A0P4Y4V0 A0A1B6CTI6 A0A0P5UAZ3 A0A0P6GJR3 A0A0N8DE38 A0A0P5Y055 A0A0P5X813 A0A0P5XUP8 A0A0P5YNS9 A0A162QRA1 A0A0P5XL26 A0A0P5Y2L0 A0A0N8A576 A0A0P5WP50 A0A0P5MXS5 A0A0P5BMC7 A0A0P5V0B5 A0A0P5Z7Y2 A0A0P6E3A7 A0A0P5X120 A0A0P6HUV1 A0A0P5E3J3 A0A195ECY0 A0A0P6EID0 A0A0P5UYZ4 A0A158NXS0 A0A0P5KKG2 A0A139WEE1 A0A151WMN1 A0A182PGK5 A0A210PEG6 A0A0P5I7D4 A0A0P5E440 A0A0P5CYJ3 A0A0P4YDK7 A0A0P5X901 A0A0P5UNV0 A0A0P5W3J8 A0A1Z5LER5 A0A0P5TR24 A0A0P4Z558 A0A0P5MS98 A0A0P5TRY0 B3P8F6 B3LY63 A0A0P6EF37 A0A293LVV1 A0A0P5UUQ5 Q7PXX9 K7IX35
A0A2H1VUS0 A0A1W1EGM7 A0A1B6LUZ4 A0A1B6IGC1 A0A1B6K1N7 A0A3S2NLJ3 A0A1B6FTD6 A0A2A4K975 J9K5F8 A0A2J7R6I8 A0A1B6J3H7 A0A2J7R6J3 A0A2H1VST3 A0A2H1X380 A0A2J7R6J1 A0A2H1VG50 A0A0P5UEK4 A0A2A4K8P8 A0A212F254 A0A0P5UKE0 A0A0N8CP26 A0A0N1IF15 A0A0P5V259 A0A0P5V115 A0A067QM95 A0A0P5VAG5 A0A154P4F4 A0A0P5D4A0 A0A0N8BXP2 A0A0N8C3M9 A0A0P6HWY3 A0A2A3EMA9 A0A0L7QMS7 A0A0P6CTA3 A0A212F293 A0A0C9QXG4 A0A0P5BCG5 H9JUB7 A0A0P5UX07 A0A0P5YE76 A0A088APM4 A0A0P5BE18 T1JDD3 I4DLH0 A0A0P4Y4V0 A0A1B6CTI6 A0A0P5UAZ3 A0A0P6GJR3 A0A0N8DE38 A0A0P5Y055 A0A0P5X813 A0A0P5XUP8 A0A0P5YNS9 A0A162QRA1 A0A0P5XL26 A0A0P5Y2L0 A0A0N8A576 A0A0P5WP50 A0A0P5MXS5 A0A0P5BMC7 A0A0P5V0B5 A0A0P5Z7Y2 A0A0P6E3A7 A0A0P5X120 A0A0P6HUV1 A0A0P5E3J3 A0A195ECY0 A0A0P6EID0 A0A0P5UYZ4 A0A158NXS0 A0A0P5KKG2 A0A139WEE1 A0A151WMN1 A0A182PGK5 A0A210PEG6 A0A0P5I7D4 A0A0P5E440 A0A0P5CYJ3 A0A0P4YDK7 A0A0P5X901 A0A0P5UNV0 A0A0P5W3J8 A0A1Z5LER5 A0A0P5TR24 A0A0P4Z558 A0A0P5MS98 A0A0P5TRY0 B3P8F6 B3LY63 A0A0P6EF37 A0A293LVV1 A0A0P5UUQ5 Q7PXX9 K7IX35
EC Number
3.2.1.45
Pubmed
EMBL
KQ459439
KPJ00977.1
AGBW02006772
OWR55189.1
KQ459602
KPI93190.1
+ More
BABH01038539 BABH01038540 BABH01038541 AGBW02010783 OWR47794.1 KQ460397 KPJ15278.1 ODYU01004571 SOQ44585.1 LT635665 SGZ49395.1 GEBQ01024969 GEBQ01012479 JAT15008.1 JAT27498.1 GECU01021735 JAS85971.1 GECU01002337 JAT05370.1 RSAL01000008 RVE53974.1 GECZ01016297 JAS53472.1 NWSH01000035 PCG80448.1 ABLF02017156 ABLF02017157 ABLF02017158 ABLF02017160 ABLF02043031 ABLF02044170 NEVH01006756 PNF36451.1 GECU01013983 JAS93723.1 PNF36457.1 ODYU01004232 SOQ43893.1 ODYU01013051 SOQ59702.1 PNF36452.1 ODYU01002190 SOQ39362.1 GDIP01114152 JAL89562.1 PCG80449.1 OWR47828.1 GDIP01111638 JAL92076.1 GDIP01112839 JAL90875.1 KQ461178 KPJ07773.1 GDIP01105549 JAL98165.1 GDIP01105548 JAL98166.1 KK853153 KDR10552.1 GDIP01102691 JAM01024.1 KQ434814 KZC06829.1 GDIP01163230 LRGB01000311 JAJ60172.1 KZS19886.1 GDIQ01134723 JAL17003.1 GDIQ01118027 JAL33699.1 GDIQ01028607 JAN66130.1 KZ288219 PBC32336.1 KQ414868 KOC59952.1 GDIP01009481 JAM94234.1 OWR47849.1 GBYB01005352 JAG75119.1 GDIP01186374 JAJ37028.1 BABH01038531 BABH01038532 BABH01038533 BABH01038534 GDIP01107443 JAL96271.1 GDIP01073106 JAM30609.1 GDIP01190080 JAJ33322.1 JH432104 AK402138 BAM18760.1 GDIP01234780 JAI88621.1 GEDC01023766 GEDC01020570 GEDC01011572 GEDC01003214 JAS13532.1 JAS16728.1 JAS25726.1 JAS34084.1 GDIP01115596 JAL88118.1 GDIQ01034555 JAN60182.1 GDIP01042743 JAM60972.1 GDIP01065954 JAM37761.1 GDIP01077038 JAM26677.1 GDIP01067454 JAM36261.1 GDIP01056052 JAM47663.1 KZS19887.1 GDIP01070494 JAM33221.1 GDIP01077037 JAM26678.1 GDIP01181329 JAJ42073.1 GDIP01083544 JAM20171.1 GDIQ01149727 JAL01999.1 GDIP01182978 JAJ40424.1 GDIP01107444 JAL96270.1 GDIP01060297 JAM43418.1 GDIQ01068405 JAN26332.1 GDIP01079198 JAM24517.1 GDIQ01038503 JAN56234.1 GDIP01147339 JAJ76063.1 KQ979074 KYN23063.1 GDIQ01076099 JAN18638.1 GDIP01107445 JAL96269.1 ADTU01003452 ADTU01003453 ADTU01003454 ADTU01003455 GDIQ01183956 JAK67769.1 KQ971354 KYB26348.1 KQ982934 KYQ49129.1 NEDP02076748 OWF34883.1 GDIQ01222210 JAK29515.1 GDIP01153134 JAJ70268.1 GDIP01167885 JAJ55517.1 GDIP01231735 JAI91666.1 GDIP01075597 JAM28118.1 GDIP01111697 JAL92017.1 GDIP01091491 LRGB01002227 JAM12224.1 KZS08348.1 GFJQ02001071 JAW05899.1 GDIP01125091 JAL78623.1 GDIP01218434 JAJ04968.1 GDIQ01152677 JAK99048.1 GDIP01122885 JAL80829.1 CH954182 EDV53980.1 CH902617 EDV43967.1 KPU80566.1 GDIQ01076098 JAN18639.1 GFWV01007904 MAA32634.1 GDIP01125092 JAL78622.1 AAAB01008987 EAA00933.5 AAZX01006377 AAZX01006727
BABH01038539 BABH01038540 BABH01038541 AGBW02010783 OWR47794.1 KQ460397 KPJ15278.1 ODYU01004571 SOQ44585.1 LT635665 SGZ49395.1 GEBQ01024969 GEBQ01012479 JAT15008.1 JAT27498.1 GECU01021735 JAS85971.1 GECU01002337 JAT05370.1 RSAL01000008 RVE53974.1 GECZ01016297 JAS53472.1 NWSH01000035 PCG80448.1 ABLF02017156 ABLF02017157 ABLF02017158 ABLF02017160 ABLF02043031 ABLF02044170 NEVH01006756 PNF36451.1 GECU01013983 JAS93723.1 PNF36457.1 ODYU01004232 SOQ43893.1 ODYU01013051 SOQ59702.1 PNF36452.1 ODYU01002190 SOQ39362.1 GDIP01114152 JAL89562.1 PCG80449.1 OWR47828.1 GDIP01111638 JAL92076.1 GDIP01112839 JAL90875.1 KQ461178 KPJ07773.1 GDIP01105549 JAL98165.1 GDIP01105548 JAL98166.1 KK853153 KDR10552.1 GDIP01102691 JAM01024.1 KQ434814 KZC06829.1 GDIP01163230 LRGB01000311 JAJ60172.1 KZS19886.1 GDIQ01134723 JAL17003.1 GDIQ01118027 JAL33699.1 GDIQ01028607 JAN66130.1 KZ288219 PBC32336.1 KQ414868 KOC59952.1 GDIP01009481 JAM94234.1 OWR47849.1 GBYB01005352 JAG75119.1 GDIP01186374 JAJ37028.1 BABH01038531 BABH01038532 BABH01038533 BABH01038534 GDIP01107443 JAL96271.1 GDIP01073106 JAM30609.1 GDIP01190080 JAJ33322.1 JH432104 AK402138 BAM18760.1 GDIP01234780 JAI88621.1 GEDC01023766 GEDC01020570 GEDC01011572 GEDC01003214 JAS13532.1 JAS16728.1 JAS25726.1 JAS34084.1 GDIP01115596 JAL88118.1 GDIQ01034555 JAN60182.1 GDIP01042743 JAM60972.1 GDIP01065954 JAM37761.1 GDIP01077038 JAM26677.1 GDIP01067454 JAM36261.1 GDIP01056052 JAM47663.1 KZS19887.1 GDIP01070494 JAM33221.1 GDIP01077037 JAM26678.1 GDIP01181329 JAJ42073.1 GDIP01083544 JAM20171.1 GDIQ01149727 JAL01999.1 GDIP01182978 JAJ40424.1 GDIP01107444 JAL96270.1 GDIP01060297 JAM43418.1 GDIQ01068405 JAN26332.1 GDIP01079198 JAM24517.1 GDIQ01038503 JAN56234.1 GDIP01147339 JAJ76063.1 KQ979074 KYN23063.1 GDIQ01076099 JAN18638.1 GDIP01107445 JAL96269.1 ADTU01003452 ADTU01003453 ADTU01003454 ADTU01003455 GDIQ01183956 JAK67769.1 KQ971354 KYB26348.1 KQ982934 KYQ49129.1 NEDP02076748 OWF34883.1 GDIQ01222210 JAK29515.1 GDIP01153134 JAJ70268.1 GDIP01167885 JAJ55517.1 GDIP01231735 JAI91666.1 GDIP01075597 JAM28118.1 GDIP01111697 JAL92017.1 GDIP01091491 LRGB01002227 JAM12224.1 KZS08348.1 GFJQ02001071 JAW05899.1 GDIP01125091 JAL78623.1 GDIP01218434 JAJ04968.1 GDIQ01152677 JAK99048.1 GDIP01122885 JAL80829.1 CH954182 EDV53980.1 CH902617 EDV43967.1 KPU80566.1 GDIQ01076098 JAN18639.1 GFWV01007904 MAA32634.1 GDIP01125092 JAL78622.1 AAAB01008987 EAA00933.5 AAZX01006377 AAZX01006727
Proteomes
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
A0A194Q614
A0A212FNA8
A0A194PPY1
H9JUB6
A0A212F248
A0A194RBR5
+ More
A0A2H1VUS0 A0A1W1EGM7 A0A1B6LUZ4 A0A1B6IGC1 A0A1B6K1N7 A0A3S2NLJ3 A0A1B6FTD6 A0A2A4K975 J9K5F8 A0A2J7R6I8 A0A1B6J3H7 A0A2J7R6J3 A0A2H1VST3 A0A2H1X380 A0A2J7R6J1 A0A2H1VG50 A0A0P5UEK4 A0A2A4K8P8 A0A212F254 A0A0P5UKE0 A0A0N8CP26 A0A0N1IF15 A0A0P5V259 A0A0P5V115 A0A067QM95 A0A0P5VAG5 A0A154P4F4 A0A0P5D4A0 A0A0N8BXP2 A0A0N8C3M9 A0A0P6HWY3 A0A2A3EMA9 A0A0L7QMS7 A0A0P6CTA3 A0A212F293 A0A0C9QXG4 A0A0P5BCG5 H9JUB7 A0A0P5UX07 A0A0P5YE76 A0A088APM4 A0A0P5BE18 T1JDD3 I4DLH0 A0A0P4Y4V0 A0A1B6CTI6 A0A0P5UAZ3 A0A0P6GJR3 A0A0N8DE38 A0A0P5Y055 A0A0P5X813 A0A0P5XUP8 A0A0P5YNS9 A0A162QRA1 A0A0P5XL26 A0A0P5Y2L0 A0A0N8A576 A0A0P5WP50 A0A0P5MXS5 A0A0P5BMC7 A0A0P5V0B5 A0A0P5Z7Y2 A0A0P6E3A7 A0A0P5X120 A0A0P6HUV1 A0A0P5E3J3 A0A195ECY0 A0A0P6EID0 A0A0P5UYZ4 A0A158NXS0 A0A0P5KKG2 A0A139WEE1 A0A151WMN1 A0A182PGK5 A0A210PEG6 A0A0P5I7D4 A0A0P5E440 A0A0P5CYJ3 A0A0P4YDK7 A0A0P5X901 A0A0P5UNV0 A0A0P5W3J8 A0A1Z5LER5 A0A0P5TR24 A0A0P4Z558 A0A0P5MS98 A0A0P5TRY0 B3P8F6 B3LY63 A0A0P6EF37 A0A293LVV1 A0A0P5UUQ5 Q7PXX9 K7IX35
A0A2H1VUS0 A0A1W1EGM7 A0A1B6LUZ4 A0A1B6IGC1 A0A1B6K1N7 A0A3S2NLJ3 A0A1B6FTD6 A0A2A4K975 J9K5F8 A0A2J7R6I8 A0A1B6J3H7 A0A2J7R6J3 A0A2H1VST3 A0A2H1X380 A0A2J7R6J1 A0A2H1VG50 A0A0P5UEK4 A0A2A4K8P8 A0A212F254 A0A0P5UKE0 A0A0N8CP26 A0A0N1IF15 A0A0P5V259 A0A0P5V115 A0A067QM95 A0A0P5VAG5 A0A154P4F4 A0A0P5D4A0 A0A0N8BXP2 A0A0N8C3M9 A0A0P6HWY3 A0A2A3EMA9 A0A0L7QMS7 A0A0P6CTA3 A0A212F293 A0A0C9QXG4 A0A0P5BCG5 H9JUB7 A0A0P5UX07 A0A0P5YE76 A0A088APM4 A0A0P5BE18 T1JDD3 I4DLH0 A0A0P4Y4V0 A0A1B6CTI6 A0A0P5UAZ3 A0A0P6GJR3 A0A0N8DE38 A0A0P5Y055 A0A0P5X813 A0A0P5XUP8 A0A0P5YNS9 A0A162QRA1 A0A0P5XL26 A0A0P5Y2L0 A0A0N8A576 A0A0P5WP50 A0A0P5MXS5 A0A0P5BMC7 A0A0P5V0B5 A0A0P5Z7Y2 A0A0P6E3A7 A0A0P5X120 A0A0P6HUV1 A0A0P5E3J3 A0A195ECY0 A0A0P6EID0 A0A0P5UYZ4 A0A158NXS0 A0A0P5KKG2 A0A139WEE1 A0A151WMN1 A0A182PGK5 A0A210PEG6 A0A0P5I7D4 A0A0P5E440 A0A0P5CYJ3 A0A0P4YDK7 A0A0P5X901 A0A0P5UNV0 A0A0P5W3J8 A0A1Z5LER5 A0A0P5TR24 A0A0P4Z558 A0A0P5MS98 A0A0P5TRY0 B3P8F6 B3LY63 A0A0P6EF37 A0A293LVV1 A0A0P5UUQ5 Q7PXX9 K7IX35
PDB
5LVX
E-value=8.72917e-65,
Score=626
Ontologies
PATHWAY
GO
PANTHER
Topology
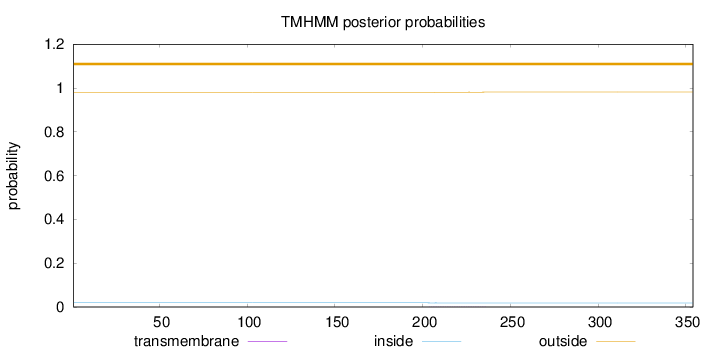
Length:
354
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01646
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.01890
outside
1 - 354
Population Genetic Test Statistics
Pi
29.911693
Theta
30.152616
Tajima's D
0.09441
CLR
0.42138
CSRT
0.390980450977451
Interpretation
Uncertain
