Gene
KWMTBOMO08028
Pre Gene Modal
BGIBMGA001129
Annotation
PREDICTED:_glucosylceramidase-like_[Bombyx_mori]
Full name
Glucosylceramidase
+ More
Putative glucosylceramidase 3
Putative glucosylceramidase 3
Location in the cell
Extracellular Reliability : 2.104
Sequence
CDS
ATGGTTTTACCATTGATACCTTTCTGTTTAAGTCTTGAAGATTTAAACAACTACGTAGTGGGTTGGATCGATTGGAACCTCTGTTTGGATCCCAACGGTGGCCCAAACTGGGCTAGCAACTTCGTAGACTCCCCAATAATTGTATATGAAGATAAAGGAGAATTCGTGAAGCAACCAATGTATTATGCATTGGGGCATTTTTCTAAATTTATTCCAAGAGGCTCAGCGCGTCTCCAAGTAACAACGCTTAGTACAGAAGGCATAGAGAACGTTGCAGTTATAACGCCGAAGGGGAATGTTGTGGTTGTTATGCAAAACAGGAAGCCACATACAGTACCTGTGAAGATATATGTGAGCAGTAGAAAAGCTATAAACATTAATATGGAACCTGAATCAATAAAAACTGTAGAAATTAATTCTGTCTCATGA
Protein
MVLPLIPFCLSLEDLNNYVVGWIDWNLCLDPNGGPNWASNFVDSPIIVYEDKGEFVKQPMYYALGHFSKFIPRGSARLQVTTLSTEGIENVAVITPKGNVVVVMQNRKPHTVPVKIYVSSRKAININMEPESIKTVEINSVS
Summary
Catalytic Activity
a beta-D-glucosyl-(1<->1')-N-acylsphing-4-enine + H2O = an N-acylsphing-4-enine + D-glucose
Similarity
Belongs to the glycosyl hydrolase 30 family.
Keywords
Alternative splicing
Complete proteome
Hydrolase
Lipid metabolism
Reference proteome
Signal
Sphingolipid metabolism
Feature
chain Glucosylceramidase
splice variant In isoform d.
splice variant In isoform d.
Uniprot
A0A2H1VA54
A0A194Q614
A0A194RBR5
A0A212F349
A0A212EPK9
H9JUB6
+ More
A0A2S2P4H2 A0A1W1EGM7 J9L5B0 A0A0N1IF15 A0A0N1INL2 A0A067QM95 A0A2A4K8P8 A0A2P8YXD8 A0A329T258 A0A2J7R6J1 G4ZEC3 A0A368H792 A0A081ANK6 W2JGX1 A0A3R7HCU5 W2NS50 W2JGV9 W2XHH3 W2QGT2 W2ZQX7 W2H8M4 W2H8L5 V9FMS3 A0A0W8D288 W4Z0W5 A0A212F254 G5A8P3 A0A0M3HW45 A0A329T209 A0A329T1T2 H3GW35 A0A1Y3B8N5 H3GW34 G4ZEB8 V9LBE0 A0A225WKD5 W2QN93 A0A329SI21 V9FRQ0 W2XM40 A0A081AVG3 W2LSY8 A0A0W8D122 W2ZX75 H3G7N0 W4YPC5 G4ZEB9 A0A0P5P886 A0A023GIV4 A0A0N7L536 A0A2H1VUS0 A0A182EXL9 A0A261A234 S4RMS8 W5ULT4 A0A2D0R7B2 A0A260ZP45 D0N0V3 G5ECR8-4 H9JUB7 F1L2Y4 A0A0P5L6K4 A0A3P6NZ05 M3VVL7 A0A183CWR4 A0A1I7V4N9 A0A2G9U3J6 A0A337SNL3 A0A225WW95 W2JP47 A0A3F2RCM9 A0A0D6M7G3 A0A0B1TDP7 A0A3R7H2D6 A0A0P5GMD5 A0A2G9U5T9 W2HHH0 A0A0P5BVB7 A0A0P5BV77 A0A2P4WZX2 A0A0C9QXG4 A0A162QRA1 D0P1U3 A0A2H1VG50 A0A0P5E3J3 V4B2P4 U3DTR8 A0A0P5WGR1 F7E7C1
A0A2S2P4H2 A0A1W1EGM7 J9L5B0 A0A0N1IF15 A0A0N1INL2 A0A067QM95 A0A2A4K8P8 A0A2P8YXD8 A0A329T258 A0A2J7R6J1 G4ZEC3 A0A368H792 A0A081ANK6 W2JGX1 A0A3R7HCU5 W2NS50 W2JGV9 W2XHH3 W2QGT2 W2ZQX7 W2H8M4 W2H8L5 V9FMS3 A0A0W8D288 W4Z0W5 A0A212F254 G5A8P3 A0A0M3HW45 A0A329T209 A0A329T1T2 H3GW35 A0A1Y3B8N5 H3GW34 G4ZEB8 V9LBE0 A0A225WKD5 W2QN93 A0A329SI21 V9FRQ0 W2XM40 A0A081AVG3 W2LSY8 A0A0W8D122 W2ZX75 H3G7N0 W4YPC5 G4ZEB9 A0A0P5P886 A0A023GIV4 A0A0N7L536 A0A2H1VUS0 A0A182EXL9 A0A261A234 S4RMS8 W5ULT4 A0A2D0R7B2 A0A260ZP45 D0N0V3 G5ECR8-4 H9JUB7 F1L2Y4 A0A0P5L6K4 A0A3P6NZ05 M3VVL7 A0A183CWR4 A0A1I7V4N9 A0A2G9U3J6 A0A337SNL3 A0A225WW95 W2JP47 A0A3F2RCM9 A0A0D6M7G3 A0A0B1TDP7 A0A3R7H2D6 A0A0P5GMD5 A0A2G9U5T9 W2HHH0 A0A0P5BVB7 A0A0P5BV77 A0A2P4WZX2 A0A0C9QXG4 A0A162QRA1 D0P1U3 A0A2H1VG50 A0A0P5E3J3 V4B2P4 U3DTR8 A0A0P5WGR1 F7E7C1
EC Number
3.2.1.45
Pubmed
EMBL
ODYU01001475
SOQ37718.1
KQ459439
KPJ00977.1
KQ460397
KPJ15278.1
+ More
AGBW02010629 OWR48154.1 AGBW02013448 OWR43423.1 BABH01038539 BABH01038540 BABH01038541 GGMR01011734 MBY24353.1 LT635665 SGZ49395.1 ABLF02008893 KQ461178 KPJ07773.1 KPJ07772.1 KK853153 KDR10552.1 NWSH01000035 PCG80449.1 PYGN01000302 PSN48919.1 MJFZ01000012 RAW42724.1 NEVH01006756 PNF36452.1 JH159154 EGZ17886.1 JOJR01000007 RCN52483.1 ANJA01000988 ETO80467.1 KI671854 ETL45017.1 MAYM02001277 MBDN02000653 RLN21147.1 RLN73856.1 KI678593 KI691839 ETL98166.1 ETM51335.1 KI671855 ETL45007.1 ANIX01001073 ETP21414.1 KI669577 ETN12367.1 ANIY01001102 ETP49406.1 KI685308 ETK91607.1 KI685309 ETK91597.1 ANIZ01000889 ETI51717.1 LNFO01001539 KUF90525.1 AAGJ04103183 AAGJ04103184 AGBW02010783 OWR47828.1 JH159161 EGZ08269.1 RAW42719.1 RAW42721.1 DS566059 MUJZ01033843 OTF77212.1 EGZ17881.1 JW876614 AFP09131.1 NBNE01000628 OWZ18186.1 KI669572 ETN14662.1 MJFZ01000176 RAW35212.1 ANIZ01000561 ETI54139.1 ANIX01000668 ETP23960.1 ANJA01000623 ETO82874.1 KI678010 KI691249 ETM00502.1 ETM53704.1 LNFP01001456 LNFO01001593 KUF86242.1 KUF90004.1 ANIY01000684 ETP51967.1 DS566004 AAGJ04003827 EGZ17882.1 GDIQ01138535 JAL13191.1 GBBM01001526 JAC33892.1 CCYD01000482 CEG40379.1 ODYU01004571 SOQ44585.1 UYRW01012587 VDN00299.1 NMWX01000014 OZF92071.1 JT411533 JT418303 AHH42685.1 NIPN01000376 NIPN01000375 OZF87457.1 OZF87500.1 DS028122 EEY67266.1 BX284604 AL031254 BABH01038531 BABH01038532 BABH01038533 BABH01038534 JI170344 ADY44488.1 GDIQ01199303 JAK52422.1 UYRT01000954 VDK28999.1 AANG04002876 KZ350247 PIO64080.1 NBNE01000176 OWZ21926.1 KI671251 ETL47408.1 MBAD02001844 MBDO02000659 RLN51536.1 RLN53096.1 KE124814 EPB78356.1 KN550413 KHJ94226.1 RLN51535.1 GDIQ01261484 JAJ90240.1 KZ349327 PIO65072.1 KI684753 ETK94015.1 GDIP01179428 JAJ43974.1 GDIP01179427 JAJ43975.1 NCKW01020142 POM58843.1 GBYB01005352 JAG75119.1 LRGB01000311 KZS19887.1 DS028257 EEY55076.1 ODYU01002190 SOQ39362.1 GDIP01147339 JAJ76063.1 KB200701 ESP00722.1 GAMT01003193 GAMS01007908 GAMQ01006239 JAB08668.1 JAB15228.1 JAB35612.1 GDIP01099492 JAM04223.1
AGBW02010629 OWR48154.1 AGBW02013448 OWR43423.1 BABH01038539 BABH01038540 BABH01038541 GGMR01011734 MBY24353.1 LT635665 SGZ49395.1 ABLF02008893 KQ461178 KPJ07773.1 KPJ07772.1 KK853153 KDR10552.1 NWSH01000035 PCG80449.1 PYGN01000302 PSN48919.1 MJFZ01000012 RAW42724.1 NEVH01006756 PNF36452.1 JH159154 EGZ17886.1 JOJR01000007 RCN52483.1 ANJA01000988 ETO80467.1 KI671854 ETL45017.1 MAYM02001277 MBDN02000653 RLN21147.1 RLN73856.1 KI678593 KI691839 ETL98166.1 ETM51335.1 KI671855 ETL45007.1 ANIX01001073 ETP21414.1 KI669577 ETN12367.1 ANIY01001102 ETP49406.1 KI685308 ETK91607.1 KI685309 ETK91597.1 ANIZ01000889 ETI51717.1 LNFO01001539 KUF90525.1 AAGJ04103183 AAGJ04103184 AGBW02010783 OWR47828.1 JH159161 EGZ08269.1 RAW42719.1 RAW42721.1 DS566059 MUJZ01033843 OTF77212.1 EGZ17881.1 JW876614 AFP09131.1 NBNE01000628 OWZ18186.1 KI669572 ETN14662.1 MJFZ01000176 RAW35212.1 ANIZ01000561 ETI54139.1 ANIX01000668 ETP23960.1 ANJA01000623 ETO82874.1 KI678010 KI691249 ETM00502.1 ETM53704.1 LNFP01001456 LNFO01001593 KUF86242.1 KUF90004.1 ANIY01000684 ETP51967.1 DS566004 AAGJ04003827 EGZ17882.1 GDIQ01138535 JAL13191.1 GBBM01001526 JAC33892.1 CCYD01000482 CEG40379.1 ODYU01004571 SOQ44585.1 UYRW01012587 VDN00299.1 NMWX01000014 OZF92071.1 JT411533 JT418303 AHH42685.1 NIPN01000376 NIPN01000375 OZF87457.1 OZF87500.1 DS028122 EEY67266.1 BX284604 AL031254 BABH01038531 BABH01038532 BABH01038533 BABH01038534 JI170344 ADY44488.1 GDIQ01199303 JAK52422.1 UYRT01000954 VDK28999.1 AANG04002876 KZ350247 PIO64080.1 NBNE01000176 OWZ21926.1 KI671251 ETL47408.1 MBAD02001844 MBDO02000659 RLN51536.1 RLN53096.1 KE124814 EPB78356.1 KN550413 KHJ94226.1 RLN51535.1 GDIQ01261484 JAJ90240.1 KZ349327 PIO65072.1 KI684753 ETK94015.1 GDIP01179428 JAJ43974.1 GDIP01179427 JAJ43975.1 NCKW01020142 POM58843.1 GBYB01005352 JAG75119.1 LRGB01000311 KZS19887.1 DS028257 EEY55076.1 ODYU01002190 SOQ39362.1 GDIP01147339 JAJ76063.1 KB200701 ESP00722.1 GAMT01003193 GAMS01007908 GAMQ01006239 JAB08668.1 JAB15228.1 JAB35612.1 GDIP01099492 JAM04223.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000005204
UP000007819
UP000027135
+ More
UP000218220 UP000245037 UP000251314 UP000235965 UP000002640 UP000252519 UP000028582 UP000053864 UP000285624 UP000285883 UP000054423 UP000054532 UP000018958 UP000018817 UP000018948 UP000053236 UP000018721 UP000052943 UP000007110 UP000036681 UP000005238 UP000198211 UP000054928 UP000077448 UP000271087 UP000216624 UP000245300 UP000221080 UP000216463 UP000006643 UP000001940 UP000011712 UP000050760 UP000095282 UP000277300 UP000284657 UP000076858 UP000030746 UP000008225
UP000218220 UP000245037 UP000251314 UP000235965 UP000002640 UP000252519 UP000028582 UP000053864 UP000285624 UP000285883 UP000054423 UP000054532 UP000018958 UP000018817 UP000018948 UP000053236 UP000018721 UP000052943 UP000007110 UP000036681 UP000005238 UP000198211 UP000054928 UP000077448 UP000271087 UP000216624 UP000245300 UP000221080 UP000216463 UP000006643 UP000001940 UP000011712 UP000050760 UP000095282 UP000277300 UP000284657 UP000076858 UP000030746 UP000008225
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
A0A2H1VA54
A0A194Q614
A0A194RBR5
A0A212F349
A0A212EPK9
H9JUB6
+ More
A0A2S2P4H2 A0A1W1EGM7 J9L5B0 A0A0N1IF15 A0A0N1INL2 A0A067QM95 A0A2A4K8P8 A0A2P8YXD8 A0A329T258 A0A2J7R6J1 G4ZEC3 A0A368H792 A0A081ANK6 W2JGX1 A0A3R7HCU5 W2NS50 W2JGV9 W2XHH3 W2QGT2 W2ZQX7 W2H8M4 W2H8L5 V9FMS3 A0A0W8D288 W4Z0W5 A0A212F254 G5A8P3 A0A0M3HW45 A0A329T209 A0A329T1T2 H3GW35 A0A1Y3B8N5 H3GW34 G4ZEB8 V9LBE0 A0A225WKD5 W2QN93 A0A329SI21 V9FRQ0 W2XM40 A0A081AVG3 W2LSY8 A0A0W8D122 W2ZX75 H3G7N0 W4YPC5 G4ZEB9 A0A0P5P886 A0A023GIV4 A0A0N7L536 A0A2H1VUS0 A0A182EXL9 A0A261A234 S4RMS8 W5ULT4 A0A2D0R7B2 A0A260ZP45 D0N0V3 G5ECR8-4 H9JUB7 F1L2Y4 A0A0P5L6K4 A0A3P6NZ05 M3VVL7 A0A183CWR4 A0A1I7V4N9 A0A2G9U3J6 A0A337SNL3 A0A225WW95 W2JP47 A0A3F2RCM9 A0A0D6M7G3 A0A0B1TDP7 A0A3R7H2D6 A0A0P5GMD5 A0A2G9U5T9 W2HHH0 A0A0P5BVB7 A0A0P5BV77 A0A2P4WZX2 A0A0C9QXG4 A0A162QRA1 D0P1U3 A0A2H1VG50 A0A0P5E3J3 V4B2P4 U3DTR8 A0A0P5WGR1 F7E7C1
A0A2S2P4H2 A0A1W1EGM7 J9L5B0 A0A0N1IF15 A0A0N1INL2 A0A067QM95 A0A2A4K8P8 A0A2P8YXD8 A0A329T258 A0A2J7R6J1 G4ZEC3 A0A368H792 A0A081ANK6 W2JGX1 A0A3R7HCU5 W2NS50 W2JGV9 W2XHH3 W2QGT2 W2ZQX7 W2H8M4 W2H8L5 V9FMS3 A0A0W8D288 W4Z0W5 A0A212F254 G5A8P3 A0A0M3HW45 A0A329T209 A0A329T1T2 H3GW35 A0A1Y3B8N5 H3GW34 G4ZEB8 V9LBE0 A0A225WKD5 W2QN93 A0A329SI21 V9FRQ0 W2XM40 A0A081AVG3 W2LSY8 A0A0W8D122 W2ZX75 H3G7N0 W4YPC5 G4ZEB9 A0A0P5P886 A0A023GIV4 A0A0N7L536 A0A2H1VUS0 A0A182EXL9 A0A261A234 S4RMS8 W5ULT4 A0A2D0R7B2 A0A260ZP45 D0N0V3 G5ECR8-4 H9JUB7 F1L2Y4 A0A0P5L6K4 A0A3P6NZ05 M3VVL7 A0A183CWR4 A0A1I7V4N9 A0A2G9U3J6 A0A337SNL3 A0A225WW95 W2JP47 A0A3F2RCM9 A0A0D6M7G3 A0A0B1TDP7 A0A3R7H2D6 A0A0P5GMD5 A0A2G9U5T9 W2HHH0 A0A0P5BVB7 A0A0P5BV77 A0A2P4WZX2 A0A0C9QXG4 A0A162QRA1 D0P1U3 A0A2H1VG50 A0A0P5E3J3 V4B2P4 U3DTR8 A0A0P5WGR1 F7E7C1
PDB
2XWE
E-value=5.35445e-28,
Score=303
Ontologies
GO
GO:0006665
GO:0004348
GO:0016787
GO:0006680
GO:0016021
GO:0036268
GO:0030278
GO:0070050
GO:0005623
GO:0032436
GO:0032715
GO:0005615
GO:0009267
GO:0043202
GO:0030259
GO:0071356
GO:0005124
GO:1904457
GO:0046513
GO:1904925
GO:0008203
GO:0050295
GO:0005783
GO:0046512
GO:0032463
GO:0007040
GO:0043243
GO:0005765
GO:1905037
GO:1903061
GO:0023021
GO:0005802
GO:0046527
GO:0035307
GO:0032006
GO:0043407
GO:1901215
GO:0005975
GO:0008236
GO:0016020
GO:0005524
GO:0006412
GO:0016876
GO:0043039
PANTHER
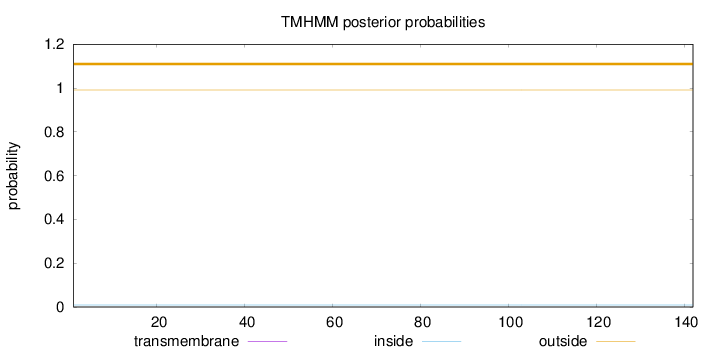
Topology
Length:
142
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00205
Exp number, first 60 AAs:
0.00065
Total prob of N-in:
0.00864
outside
1 - 142
Population Genetic Test Statistics
Pi
17.762122
Theta
17.727465
Tajima's D
0.007775
CLR
0.006249
CSRT
0.384180790960452
Interpretation
Uncertain
