Gene
KWMTBOMO08025
Pre Gene Modal
BGIBMGA001026
Annotation
PREDICTED:_glutamate_receptor_ionotropic?_delta-2_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.83
Sequence
CDS
ATGGCCGGTATCGAGTTGATCATATCTTCAATTTGCAACGCAACAATTTGTAATGCCGTGTATGATAATCCGCTAACAGATGGACATGTGTCACAAAAGCAAACCGAGACTTTACTGTTGGCGAATGAAATAAATGGGAAGCACTTAAAGATCGGCACGTATAATAATTTCCCTTTAAGCTGGATTGAAAGAGACGAAAATGGTACAGTGCAGGCGTACGGCGTGGCGTTTAAAATCATAGACATACTACAGCAAAAGTTTAACTTTACATACGAAATCGTAATACCACACAGGAACTTTGAAATCGGTGGCAGCAAACCTGAGGACTCTTTGATAGGGCTAACGAATACAAGTAAAGTAGACATGATAGCTGCGTTCATACCAAGGCTCGTTAGGTTCCGAAAATTGGTGACGTTCTCAAGAGATTTAGACGAAGGTGTTTGGATGATGATGCTTAGACGGCCAAAGGAATCCGCCGCTGGCTCTGGTCTTTTAGCGCCCTTTAATAACTTTGTATGGTACGTAACTCTTGCTTCGGTACTGTGTTATGGGCCGTGCATATGTTTCCTGACTCACGTGAGGTCAAAACTAATTAAGAATGAAGAACGGCCGCTGCGACTTTCGCCGAGTTTCTGGTTCGTGTATAGCGCTTTTATAAAGCAAAGCACAAACTTAGCTCCCGAAGCTAACACAACACGTGTTTTGTTCGCAACGTGGTGGCTATTCATTATATTGCTCTCAGCGTTTTACACGGCAAACTTGACTGCTTTCCTGACATTGTCCAAGTTCACACTAGACATTGAGACTCCGGAGGATTTGTATAAGAAAAATTACCGTTGGGTTTCTGTTGAGGGTGGTTCCGTTCAATACACTGTTAAAACACAGGACGAAGACCTTTACTATTTAAACAAAATGGTGACCAGTGGCCGAGCAGAATTTCGTACGTTGTCGCCAGACCAGGAGTATCTCCCAATTGTGAAAGCTGGGGCTGTTCTCGTCAAGGAGATGATTTCTCTAGAACATCTCATGTATGGGGACTACCTGACGAAGACTCGTGAAGGCGTCGAAGAGGCTAAACGATGCACTTACGTTGTGGCGCCCAAGCCTTTCATGAAAAAGCCCAGAGCGTTCGTGTATCCTGTTGGCAGTAAGCTGAAGAGCCTTTTTGATCCTACTCTTGCATACATATTACAATCGGGTATTATAGATTATCTTGAACACAAAGATTTGCCGTCCACCACGATATGTCCTCTGGATTTGCAGTCCAAAGACAGGCAGTTGACCAATAGTCACCTCATGATGACTTATTACATAATGTGCGTTGGCCTCGCCTCGGGTTTAGCTGTTTTTGTAGTGGAGATATTAGTCAAACGATACATCAATATTAAGATAAAACCTATCGATAAAGTGAAATTGAAGAAGTTCAAACGGAGTAAGCGCTCCCCACGTTACGACGACAGTGGACCGCCGCCATATGAATCCTTGTTTGTTAAACCGAAGTTCAAAGACTCCGAAAAACGTTGGAAGATGATCAACGGCAGGGAATATTACGTCTATGAAGATGCCCGAGGGGGCACCAGGCTTGTTCCCGTGAGGACTCCATCGGCTTTCTTAAATGATGACTCTGGCTTTGCTGACATACATGGATGA
Protein
MAGIELIISSICNATICNAVYDNPLTDGHVSQKQTETLLLANEINGKHLKIGTYNNFPLSWIERDENGTVQAYGVAFKIIDILQQKFNFTYEIVIPHRNFEIGGSKPEDSLIGLTNTSKVDMIAAFIPRLVRFRKLVTFSRDLDEGVWMMMLRRPKESAAGSGLLAPFNNFVWYVTLASVLCYGPCICFLTHVRSKLIKNEERPLRLSPSFWFVYSAFIKQSTNLAPEANTTRVLFATWWLFIILLSAFYTANLTAFLTLSKFTLDIETPEDLYKKNYRWVSVEGGSVQYTVKTQDEDLYYLNKMVTSGRAEFRTLSPDQEYLPIVKAGAVLVKEMISLEHLMYGDYLTKTREGVEEAKRCTYVVAPKPFMKKPRAFVYPVGSKLKSLFDPTLAYILQSGIIDYLEHKDLPSTTICPLDLQSKDRQLTNSHLMMTYYIMCVGLASGLAVFVVEILVKRYINIKIKPIDKVKLKKFKRSKRSPRYDDSGPPPYESLFVKPKFKDSEKRWKMINGREYYVYEDARGGTRLVPVRTPSAFLNDDSGFADIHG
Summary
Uniprot
H9IUU6
A0A3G6V6P6
A0A0F7QHP7
A0A194QC15
A0A1B3P5G3
A0A140G9H7
+ More
A0A345BF40 A0A146JYZ2 A0A0U3BFA4 A0A076E9D5 A0A0K8TUX1 A0A076E9E3 A0A2K8GKZ9 A0A386H956 A0A223HCY0 A0A3S2LA01 A0A1Q1PP68 H9A5S5 A0A223HDB6 E5FIB2 A0A2H1VA48 A0A1B3B6Z5 U5PZQ9 A0A0B4ZWK3 A0A194RCE5 A0A3Q8HDG9 A0A2K8GKY4 A0A2A4J6F4 B4KV54 B3M3J3 Q9VW78 A0A0J9RZR7 B4IA41 B4LFY5 A0A1W4W7R6 B4IYS9 B3NE31 B4PFT4 B4MKI2 Q2M090 A0A3B0JL64 B4GRY9 A0A068FBN9 A0A0M4EAZ5 A0A1A9XC85 A0A0L0CKJ5 A0A1A9VHR2 A0A1S4FD88 A0A182GSE3 A0A336K8J6 A0A1B0CLI0 A0A2P8ZI36 A0A182GRK7 A0A2P1JHH3 A0A2U9NK99 A0A182N2Z0 A0A1I8N5B2 A0A182RKF8 A0A1I8NWC1 A0A182TYZ9 A0A1B0G8W3 W8BPH8 A0A0K8UQ12 A0A182JQY4 A0A348AZZ2 A0A0G2UKD7 A0A182XJM9 A0A0C5DAS1 A0A1B3B7D1 A0A310S9V3 A0A182YBM8 A0A182NZY1 B4QRG1 A0A2M4APY3 A0A182V953 W5JH25 A0A0U5A7M1 A0A182HHQ0 A0A182W227 A0A067RKE4 A0A182FTQ3 A0A0M5KFI4 A0A2P0ZPL5 A0A0S3J2L2 B0XJ31 R9PSQ3 Q176J1 A0A182MCN5 D2A190 A0A2J7R359 A0A3L9LU07 A0A1S4H944 Q7PZL3 A0A3L8E5T9 A0A0L7R8U2 A0A154PIW4 B0WFT4 A0A2S2NIV4 A0A2S2QCS6
A0A345BF40 A0A146JYZ2 A0A0U3BFA4 A0A076E9D5 A0A0K8TUX1 A0A076E9E3 A0A2K8GKZ9 A0A386H956 A0A223HCY0 A0A3S2LA01 A0A1Q1PP68 H9A5S5 A0A223HDB6 E5FIB2 A0A2H1VA48 A0A1B3B6Z5 U5PZQ9 A0A0B4ZWK3 A0A194RCE5 A0A3Q8HDG9 A0A2K8GKY4 A0A2A4J6F4 B4KV54 B3M3J3 Q9VW78 A0A0J9RZR7 B4IA41 B4LFY5 A0A1W4W7R6 B4IYS9 B3NE31 B4PFT4 B4MKI2 Q2M090 A0A3B0JL64 B4GRY9 A0A068FBN9 A0A0M4EAZ5 A0A1A9XC85 A0A0L0CKJ5 A0A1A9VHR2 A0A1S4FD88 A0A182GSE3 A0A336K8J6 A0A1B0CLI0 A0A2P8ZI36 A0A182GRK7 A0A2P1JHH3 A0A2U9NK99 A0A182N2Z0 A0A1I8N5B2 A0A182RKF8 A0A1I8NWC1 A0A182TYZ9 A0A1B0G8W3 W8BPH8 A0A0K8UQ12 A0A182JQY4 A0A348AZZ2 A0A0G2UKD7 A0A182XJM9 A0A0C5DAS1 A0A1B3B7D1 A0A310S9V3 A0A182YBM8 A0A182NZY1 B4QRG1 A0A2M4APY3 A0A182V953 W5JH25 A0A0U5A7M1 A0A182HHQ0 A0A182W227 A0A067RKE4 A0A182FTQ3 A0A0M5KFI4 A0A2P0ZPL5 A0A0S3J2L2 B0XJ31 R9PSQ3 Q176J1 A0A182MCN5 D2A190 A0A2J7R359 A0A3L9LU07 A0A1S4H944 Q7PZL3 A0A3L8E5T9 A0A0L7R8U2 A0A154PIW4 B0WFT4 A0A2S2NIV4 A0A2S2QCS6
Pubmed
19121390
30465940
25803580
26354079
27538507
27004525
+ More
29727827 26657286 24998398 26017144 28736530 28150741 22363688 21091811 23894529 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21220098 26109357 26109356 22936249 17550304 15632085 25880816 26108605 26483478 29403074 25315136 24495485 30026095 25665775 27171401 25244985 20920257 23761445 26760975 24845553 26265180 26626891 23517120 17510324 18362917 19820115 12364791 30249741
29727827 26657286 24998398 26017144 28736530 28150741 22363688 21091811 23894529 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21220098 26109357 26109356 22936249 17550304 15632085 25880816 26108605 26483478 29403074 25315136 24495485 30026095 25665775 27171401 25244985 20920257 23761445 26760975 24845553 26265180 26626891 23517120 17510324 18362917 19820115 12364791 30249741
EMBL
BABH01000472
BABH01000473
MH193968
AZB49414.1
LC017795
BAR64809.1
+ More
KQ459439 KPJ00971.1 KX655901 AOG12850.1 KU702629 AMM70656.1 MG820693 AXF48864.1 KX084512 GEDO01000007 ARO76467.1 JAP88619.1 KP975099 ALT31618.1 KF487716 AII01114.1 GCVX01000142 JAI18088.1 KF487726 AII01124.1 KX585408 ARO70301.1 MG546649 AYD42272.1 KY283577 AST36237.1 RSAL01000074 RVE48972.1 KY325457 AQM73618.1 JN836725 AFC91765.1 KY283706 AST36365.1 HM562976 ADR64687.1 ODYU01001475 SOQ37715.1 KT588077 AOE47987.1 KC907740 AGY49253.1 KJ542756 AJD81640.1 KQ460397 KPJ15272.1 MF625614 AXY83441.1 KX585395 ARO70288.1 NWSH01002836 PCG67459.1 CH933809 EDW19394.2 CH902618 EDV39254.1 AE014296 BT022767 BT022775 HQ600589 AAF49071.1 AAY55183.1 AAY55191.1 ADU79033.1 CM002912 KMZ00685.1 CH480826 EDW44154.1 CH940647 EDW70384.1 CH916366 EDV96616.1 CH954178 EDV52455.1 CM000159 EDW95231.1 CH963847 EDW72688.1 CH379069 EAL31045.1 OUUW01000002 SPP76120.1 CH479188 EDW40524.1 KJ702127 AID61281.1 CP012525 ALC44516.1 JRES01000275 KNC32757.1 JXUM01084367 JXUM01084368 JXUM01084369 KQ563433 KXJ73839.1 UFQS01000174 UFQT01000174 SSX00737.1 SSX21117.1 AJWK01017289 PYGN01000050 PSN56156.1 JXUM01016354 JXUM01016355 JXUM01016356 KQ560456 KXJ82326.1 MG766214 AVN97884.1 MF385156 AWT23327.1 CCAG010010272 GAMC01003485 JAC03071.1 GDHF01023706 JAI28608.1 FX985983 BBD43518.1 KP743672 AKI28988.1 KP296779 AJO62243.1 KU291866 AOE48116.1 GGOB01000052 MCH29448.1 CM000363 EDX11174.1 GGFK01009471 MBW42792.1 ADMH02001207 ETN63667.1 FX982935 BAU20261.1 APCN01002432 KK852417 KDR24341.1 KP843210 ALD51347.1 KY817092 AVH87308.1 KT381534 ALR72540.1 DS233427 EDS29926.1 GABX01000011 JAA74508.1 CH477386 EAT42092.1 AXCM01000173 KQ971338 EFA01567.1 NEVH01007822 PNF35281.1 KB466529 RLZ02255.1 AAAB01008986 EAA00639.3 QOIP01000001 RLU27645.1 KQ414631 KOC67248.1 KQ434931 KZC11826.1 DS231920 EDS26428.1 GGMR01004087 MBY16706.1 GGMS01006345 MBY75548.1
KQ459439 KPJ00971.1 KX655901 AOG12850.1 KU702629 AMM70656.1 MG820693 AXF48864.1 KX084512 GEDO01000007 ARO76467.1 JAP88619.1 KP975099 ALT31618.1 KF487716 AII01114.1 GCVX01000142 JAI18088.1 KF487726 AII01124.1 KX585408 ARO70301.1 MG546649 AYD42272.1 KY283577 AST36237.1 RSAL01000074 RVE48972.1 KY325457 AQM73618.1 JN836725 AFC91765.1 KY283706 AST36365.1 HM562976 ADR64687.1 ODYU01001475 SOQ37715.1 KT588077 AOE47987.1 KC907740 AGY49253.1 KJ542756 AJD81640.1 KQ460397 KPJ15272.1 MF625614 AXY83441.1 KX585395 ARO70288.1 NWSH01002836 PCG67459.1 CH933809 EDW19394.2 CH902618 EDV39254.1 AE014296 BT022767 BT022775 HQ600589 AAF49071.1 AAY55183.1 AAY55191.1 ADU79033.1 CM002912 KMZ00685.1 CH480826 EDW44154.1 CH940647 EDW70384.1 CH916366 EDV96616.1 CH954178 EDV52455.1 CM000159 EDW95231.1 CH963847 EDW72688.1 CH379069 EAL31045.1 OUUW01000002 SPP76120.1 CH479188 EDW40524.1 KJ702127 AID61281.1 CP012525 ALC44516.1 JRES01000275 KNC32757.1 JXUM01084367 JXUM01084368 JXUM01084369 KQ563433 KXJ73839.1 UFQS01000174 UFQT01000174 SSX00737.1 SSX21117.1 AJWK01017289 PYGN01000050 PSN56156.1 JXUM01016354 JXUM01016355 JXUM01016356 KQ560456 KXJ82326.1 MG766214 AVN97884.1 MF385156 AWT23327.1 CCAG010010272 GAMC01003485 JAC03071.1 GDHF01023706 JAI28608.1 FX985983 BBD43518.1 KP743672 AKI28988.1 KP296779 AJO62243.1 KU291866 AOE48116.1 GGOB01000052 MCH29448.1 CM000363 EDX11174.1 GGFK01009471 MBW42792.1 ADMH02001207 ETN63667.1 FX982935 BAU20261.1 APCN01002432 KK852417 KDR24341.1 KP843210 ALD51347.1 KY817092 AVH87308.1 KT381534 ALR72540.1 DS233427 EDS29926.1 GABX01000011 JAA74508.1 CH477386 EAT42092.1 AXCM01000173 KQ971338 EFA01567.1 NEVH01007822 PNF35281.1 KB466529 RLZ02255.1 AAAB01008986 EAA00639.3 QOIP01000001 RLU27645.1 KQ414631 KOC67248.1 KQ434931 KZC11826.1 DS231920 EDS26428.1 GGMR01004087 MBY16706.1 GGMS01006345 MBY75548.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000053240
UP000218220
UP000009192
+ More
UP000007801 UP000000803 UP000001292 UP000008792 UP000192221 UP000001070 UP000008711 UP000002282 UP000007798 UP000001819 UP000268350 UP000008744 UP000092553 UP000092443 UP000037069 UP000078200 UP000069940 UP000249989 UP000092461 UP000245037 UP000075884 UP000095301 UP000075900 UP000095300 UP000075902 UP000092444 UP000075881 UP000076407 UP000076408 UP000075885 UP000000304 UP000075903 UP000000673 UP000075840 UP000075920 UP000027135 UP000069272 UP000002320 UP000008820 UP000075883 UP000007266 UP000235965 UP000007062 UP000279307 UP000053825 UP000076502
UP000007801 UP000000803 UP000001292 UP000008792 UP000192221 UP000001070 UP000008711 UP000002282 UP000007798 UP000001819 UP000268350 UP000008744 UP000092553 UP000092443 UP000037069 UP000078200 UP000069940 UP000249989 UP000092461 UP000245037 UP000075884 UP000095301 UP000075900 UP000095300 UP000075902 UP000092444 UP000075881 UP000076407 UP000076408 UP000075885 UP000000304 UP000075903 UP000000673 UP000075840 UP000075920 UP000027135 UP000069272 UP000002320 UP000008820 UP000075883 UP000007266 UP000235965 UP000007062 UP000279307 UP000053825 UP000076502
Interpro
SUPFAM
SSF49899
SSF49899
ProteinModelPortal
H9IUU6
A0A3G6V6P6
A0A0F7QHP7
A0A194QC15
A0A1B3P5G3
A0A140G9H7
+ More
A0A345BF40 A0A146JYZ2 A0A0U3BFA4 A0A076E9D5 A0A0K8TUX1 A0A076E9E3 A0A2K8GKZ9 A0A386H956 A0A223HCY0 A0A3S2LA01 A0A1Q1PP68 H9A5S5 A0A223HDB6 E5FIB2 A0A2H1VA48 A0A1B3B6Z5 U5PZQ9 A0A0B4ZWK3 A0A194RCE5 A0A3Q8HDG9 A0A2K8GKY4 A0A2A4J6F4 B4KV54 B3M3J3 Q9VW78 A0A0J9RZR7 B4IA41 B4LFY5 A0A1W4W7R6 B4IYS9 B3NE31 B4PFT4 B4MKI2 Q2M090 A0A3B0JL64 B4GRY9 A0A068FBN9 A0A0M4EAZ5 A0A1A9XC85 A0A0L0CKJ5 A0A1A9VHR2 A0A1S4FD88 A0A182GSE3 A0A336K8J6 A0A1B0CLI0 A0A2P8ZI36 A0A182GRK7 A0A2P1JHH3 A0A2U9NK99 A0A182N2Z0 A0A1I8N5B2 A0A182RKF8 A0A1I8NWC1 A0A182TYZ9 A0A1B0G8W3 W8BPH8 A0A0K8UQ12 A0A182JQY4 A0A348AZZ2 A0A0G2UKD7 A0A182XJM9 A0A0C5DAS1 A0A1B3B7D1 A0A310S9V3 A0A182YBM8 A0A182NZY1 B4QRG1 A0A2M4APY3 A0A182V953 W5JH25 A0A0U5A7M1 A0A182HHQ0 A0A182W227 A0A067RKE4 A0A182FTQ3 A0A0M5KFI4 A0A2P0ZPL5 A0A0S3J2L2 B0XJ31 R9PSQ3 Q176J1 A0A182MCN5 D2A190 A0A2J7R359 A0A3L9LU07 A0A1S4H944 Q7PZL3 A0A3L8E5T9 A0A0L7R8U2 A0A154PIW4 B0WFT4 A0A2S2NIV4 A0A2S2QCS6
A0A345BF40 A0A146JYZ2 A0A0U3BFA4 A0A076E9D5 A0A0K8TUX1 A0A076E9E3 A0A2K8GKZ9 A0A386H956 A0A223HCY0 A0A3S2LA01 A0A1Q1PP68 H9A5S5 A0A223HDB6 E5FIB2 A0A2H1VA48 A0A1B3B6Z5 U5PZQ9 A0A0B4ZWK3 A0A194RCE5 A0A3Q8HDG9 A0A2K8GKY4 A0A2A4J6F4 B4KV54 B3M3J3 Q9VW78 A0A0J9RZR7 B4IA41 B4LFY5 A0A1W4W7R6 B4IYS9 B3NE31 B4PFT4 B4MKI2 Q2M090 A0A3B0JL64 B4GRY9 A0A068FBN9 A0A0M4EAZ5 A0A1A9XC85 A0A0L0CKJ5 A0A1A9VHR2 A0A1S4FD88 A0A182GSE3 A0A336K8J6 A0A1B0CLI0 A0A2P8ZI36 A0A182GRK7 A0A2P1JHH3 A0A2U9NK99 A0A182N2Z0 A0A1I8N5B2 A0A182RKF8 A0A1I8NWC1 A0A182TYZ9 A0A1B0G8W3 W8BPH8 A0A0K8UQ12 A0A182JQY4 A0A348AZZ2 A0A0G2UKD7 A0A182XJM9 A0A0C5DAS1 A0A1B3B7D1 A0A310S9V3 A0A182YBM8 A0A182NZY1 B4QRG1 A0A2M4APY3 A0A182V953 W5JH25 A0A0U5A7M1 A0A182HHQ0 A0A182W227 A0A067RKE4 A0A182FTQ3 A0A0M5KFI4 A0A2P0ZPL5 A0A0S3J2L2 B0XJ31 R9PSQ3 Q176J1 A0A182MCN5 D2A190 A0A2J7R359 A0A3L9LU07 A0A1S4H944 Q7PZL3 A0A3L8E5T9 A0A0L7R8U2 A0A154PIW4 B0WFT4 A0A2S2NIV4 A0A2S2QCS6
PDB
4U5D
E-value=9.51752e-16,
Score=205
Ontologies
GO
PANTHER
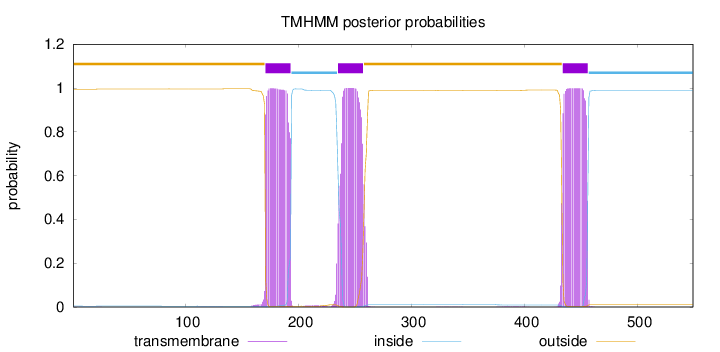
Topology
Length:
549
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
67.6124800000001
Exp number, first 60 AAs:
0.02265
Total prob of N-in:
0.00510
outside
1 - 170
TMhelix
171 - 193
inside
194 - 234
TMhelix
235 - 257
outside
258 - 433
TMhelix
434 - 456
inside
457 - 549
Population Genetic Test Statistics
Pi
1.413937
Theta
5.446336
Tajima's D
-2.216823
CLR
184.632498
CSRT
0.00409979501024949
Interpretation
Possibly Positive selection
