Gene
KWMTBOMO08021
Pre Gene Modal
BGIBMGA001024
Annotation
PREDICTED:_transmembrane_and_TPR_repeat-containing_protein_CG4050_[Plutella_xylostella]
Full name
Protein O-mannosyl-transferase TMTC3
Alternative Name
Transmembrane and TPR repeat-containing protein CG4050
Location in the cell
PlasmaMembrane Reliability : 1.668
Sequence
CDS
ATGTTCCTACCGGAGTTCGCAAGTTTCGTCGCCGCCATCTTGTTCGCCGTGCATCCGATTCACACCGAAGCGGTTACGGGTGTAGTGGGAAGAGCCGAGATGTTGTCATCGCTTTTTTTCCTCGGAGCGCTCCTTTGTTATGCCAAAGCTGCTAATAGGCGAAGAAATACAGATTGGCGCTACGTGGCTGCGTGCACTGCATGCGTGGGCATCGCAATGCTATGCAAGGAACAAGGCATTACTGTTACCGGAGTGTGTGTGGTCTACGAACTATTAGTCGCACAAAAGCGTCGAAGCGCCACCGGCGAAAGCCGCTGGTCGTGGATGGACGCCTGGGGTAAAGGCTCCGGTTGGGGTTGCGCCTGTGGCGAGGCTGCTCGAAGGGTCGCCACAGTTTGTTGCGCTACGTTAGCGTTGCTGGCCGCGAGGTTACACGTTATGGGCGCTCAGCTGCCAGTGTTCACCAGATTCGACAATCCGGCCGGAGCGGCACCTGGTCCGACTAGACACTTGACCTTCGCCTATTTACCGGCGTTGAATGCTTGGCTCTTGGCATTACCCGAAGCCTTATGCTGCGACTGGACCATGGGCACCGTAGCTCTGGTCAAATCGTGGCGAGATCCTCGGAACGTCGCCACGTTGGCGTTAGCTGCAGTTCTCGTTGCCGCGGCTATTCATGCCTTGCGTACAAGATGTTCCGCCTTATCCATGGGTCTGGCTTTGCTAGTCTTACCTTTCTTGCCAGCGTCTAACCTTTTCTTTCCCGTGGGTTTCGTGGTGGCCGAACGAGTTCTTTATATGCCTTCGATGGGTTGGTGCTTGTTGGTCGCACACGGATGGAGAATAGTCGCGCAAAGCCGAGCCAAACTGGCTGCGGCCGCTCTCATATTTCTCCTTCTGGCGTTCAGTGCCAAAACGTACATGAGAAACTGGGATTGGAAGACTGAATATTCCATCTTCGCCTCCGGGTTAAAGGTAAATCGGTTCAATGCCAAATTGTACAACAACGTTGGACACGCTTTGGAAGCCGAAGGTAAATATTCCGAAGCGTTGGACTTCTTCAACACCGCAGTGAGCGTACAACCCGATGACGTAGGGGCCCATATCAACGTGGGCCGAACTTACAACCATTTAGGCAGATACCAGGAAGCCGAAGATGCCTACGTGAAAGCGAAGTCGTTGCTACCGAAAGCGAAACCCGGTGAATCGTACCAAGCCAGGATAGCGCCCAACCATTTGAACGTCTTCCTCAATCTGGCCAACTTAATATCTAAGAACGCAACGAGGCTGGAGGAGGCTGACATGCTGTACCGACAAGCGATCAGCATGAGGGCCGATTACACTCAAGCTTATATAAATCGAGGGGATATTCTGATCAAATTGAACAGGACAAAAGAAGCGCAGGAGGTTTACGAACGCGCCCTATTGTACGACAGCGGAAACCCGGACATTTACTATAACTTGGGAGTTGTCCTTCTAGAACAAGGGAAGGCGTCCCAAGCTCTCGCGTACTTGGACAAGGCATTAGAACTTGAACCGGAACACGAGCAGGCTCTGTTGAATTCCGCCATACTCCTGCAAGAATTGGGAGCTGCTGATCTAAGGCATCTGGCCAGGCAGCGACTGCTGAAACTACTCGATAAGGACGCTACCAACGAACGCGTGCACTTCAATTTGGGCATGGTGTGTATGGACGAGGGCGACGCGGAGTGCGCCGAGCGCTGGTTCCGGGCGGCCGTTCACCTCAAGCCCGACTTCCGGTCGGCGCTATTCAACCTGGCGCTGCTGCTCGCGGACCGCCGCCGCCCCTTGGAGGCCGCCCCGTTTTTGAAACAGCTCGTCCGACACCACCCGGACCACGTGAAGGCGCTCGTACTACTCGGCGACATTTACATCAATTCCGTGAAGGACTTGGACGCGGCCGAGAGCTGCTACCGTCGCATACTGGAGTTAGAGCCGGACAACGTGCAGGCCTTGCACAACCTTTGCGTGGTGGCCGTCGAGCGCGGGAAGCTCGCAGTTGCCGAGGAGTGCCTCACCAGGGCGGCAGCGCTCGCTCCTCACGAACACTACATACAGCGACATCTCGCGGTCGTGAAAGCGAGACGGGCAGCTAACACTTCTGTCCCCTCGAAGTCGGAACCGCCCGCACCCACCGCCACCACTGAGGTGCGAGCGAGATGGAACTACATCCCGCAGCAACCCCCCGATCCACACGAGACTGACGCGTCATAG
Protein
MFLPEFASFVAAILFAVHPIHTEAVTGVVGRAEMLSSLFFLGALLCYAKAANRRRNTDWRYVAACTACVGIAMLCKEQGITVTGVCVVYELLVAQKRRSATGESRWSWMDAWGKGSGWGCACGEAARRVATVCCATLALLAARLHVMGAQLPVFTRFDNPAGAAPGPTRHLTFAYLPALNAWLLALPEALCCDWTMGTVALVKSWRDPRNVATLALAAVLVAAAIHALRTRCSALSMGLALLVLPFLPASNLFFPVGFVVAERVLYMPSMGWCLLVAHGWRIVAQSRAKLAAAALIFLLLAFSAKTYMRNWDWKTEYSIFASGLKVNRFNAKLYNNVGHALEAEGKYSEALDFFNTAVSVQPDDVGAHINVGRTYNHLGRYQEAEDAYVKAKSLLPKAKPGESYQARIAPNHLNVFLNLANLISKNATRLEEADMLYRQAISMRADYTQAYINRGDILIKLNRTKEAQEVYERALLYDSGNPDIYYNLGVVLLEQGKASQALAYLDKALELEPEHEQALLNSAILLQELGAADLRHLARQRLLKLLDKDATNERVHFNLGMVCMDEGDAECAERWFRAAVHLKPDFRSALFNLALLLADRRRPLEAAPFLKQLVRHHPDHVKALVLLGDIYINSVKDLDAAESCYRRILELEPDNVQALHNLCVVAVERGKLAVAEECLTRAAALAPHEHYIQRHLAVVKARRAANTSVPSKSEPPAPTATTEVRARWNYIPQQPPDPHETDAS
Summary
Description
Transfers mannosyl residues to the hydroxyl group of serine or threonine residues.
Catalytic Activity
dolichyl beta-D-mannosyl phosphate + L-seryl-[protein] = 3-O-(alpha-D-mannosyl)-L-seryl-[protein] + dolichyl phosphate + H(+)
dolichyl beta-D-mannosyl phosphate + L-threonyl-[protein] = 3-O-(alpha-D-mannosyl)-L-threonyl-[protein] + dolichyl phosphate + H(+)
dolichyl beta-D-mannosyl phosphate + L-threonyl-[protein] = 3-O-(alpha-D-mannosyl)-L-threonyl-[protein] + dolichyl phosphate + H(+)
Similarity
Belongs to the TMTC family.
Keywords
Complete proteome
Endoplasmic reticulum
Glycoprotein
Membrane
Reference proteome
Repeat
TPR repeat
Transferase
Transmembrane
Transmembrane helix
Feature
chain Protein O-mannosyl-transferase TMTC3
Uniprot
H9IUU4
A0A194Q7J7
A0A194RH60
A0A2H1VP12
A0A212F329
A0A1W4X7D0
+ More
A0A0T6B9B0 A0A1Y1MZA3 D1ZZM8 A0A087ZXM0 A0A1Y1MZA5 A0A151IJE7 A0A195ECP6 E2BZ92 A0A158NY64 K7IYU1 A0A195EV02 A0A195AZT2 E2AXX3 A0A026WJF4 A0A1B6CC38 A0A1B6L999 A0A0L7R8N4 A0A0C9RE72 A0A1B6F0B4 A0A1B6JSP7 A0A0V0G6Y8 T1HMF2 A0A224XJU4 E9J0P2 E0VDN7 A0A0N1IU94 A0A146L2T0 A0A0A9XE34 A0A232ESA3 A0A2A3EC51 U4UDB0 A0A1J1HF50 J9JZ49 A0A2H8TF53 A0A2S2R6V2 A0A310SKP5 A0A1Y9GLF6 A0A164PQ75 A0A2R5LEI4 A0A0N8AEJ6 V9IB44 A0A0J7NZC1 A0A2P2HX23 A0A0N8AGA7 A0A0P5ISW0 A0A0N8BLU3 A0A1D2N4K5 A0A034VHM9 A0A0P5UL86 A0A0P6IJU7 A0A147BMG7 A0A0M3QUR2 A0A224Z242 A0A0P5WAK1 B4LMR8 T1J5F2 A0A0A1WKU4 A0A3B0J1P0 A0A0J9RJ04 Q28X17 B3MDU2 B4QFM3 B3NJY1 E2QCJ5 B4P9H7 Q7K4B6 B4MKC8 B4I7K0 A0A336KWY0 A0A182QTG0 B4KNB0 W8BJJ3 A0A1A9UVB9 A0A1W4VI06 A0A1B0ACW3 A0A1B0FAZ2 A0A1A9YBE8 A0A1B0B2G1 A0A182IMN2 B4J6R6 A0A0K2TUB6 A0A084WI12 E9H3D7 A0A0K8W6E4 A0A0P5VCQ0 A0A0K8VW81 T1ITR6 A0A2M4DSF7 C3Y7I3 N6TXE2 N6UID8 A0A1S3KH78 B4H4W9
A0A0T6B9B0 A0A1Y1MZA3 D1ZZM8 A0A087ZXM0 A0A1Y1MZA5 A0A151IJE7 A0A195ECP6 E2BZ92 A0A158NY64 K7IYU1 A0A195EV02 A0A195AZT2 E2AXX3 A0A026WJF4 A0A1B6CC38 A0A1B6L999 A0A0L7R8N4 A0A0C9RE72 A0A1B6F0B4 A0A1B6JSP7 A0A0V0G6Y8 T1HMF2 A0A224XJU4 E9J0P2 E0VDN7 A0A0N1IU94 A0A146L2T0 A0A0A9XE34 A0A232ESA3 A0A2A3EC51 U4UDB0 A0A1J1HF50 J9JZ49 A0A2H8TF53 A0A2S2R6V2 A0A310SKP5 A0A1Y9GLF6 A0A164PQ75 A0A2R5LEI4 A0A0N8AEJ6 V9IB44 A0A0J7NZC1 A0A2P2HX23 A0A0N8AGA7 A0A0P5ISW0 A0A0N8BLU3 A0A1D2N4K5 A0A034VHM9 A0A0P5UL86 A0A0P6IJU7 A0A147BMG7 A0A0M3QUR2 A0A224Z242 A0A0P5WAK1 B4LMR8 T1J5F2 A0A0A1WKU4 A0A3B0J1P0 A0A0J9RJ04 Q28X17 B3MDU2 B4QFM3 B3NJY1 E2QCJ5 B4P9H7 Q7K4B6 B4MKC8 B4I7K0 A0A336KWY0 A0A182QTG0 B4KNB0 W8BJJ3 A0A1A9UVB9 A0A1W4VI06 A0A1B0ACW3 A0A1B0FAZ2 A0A1A9YBE8 A0A1B0B2G1 A0A182IMN2 B4J6R6 A0A0K2TUB6 A0A084WI12 E9H3D7 A0A0K8W6E4 A0A0P5VCQ0 A0A0K8VW81 T1ITR6 A0A2M4DSF7 C3Y7I3 N6TXE2 N6UID8 A0A1S3KH78 B4H4W9
EC Number
2.4.1.109
Pubmed
19121390
26354079
22118469
28004739
18362917
19820115
+ More
20798317 21347285 20075255 24508170 30249741 21282665 20566863 26823975 25401762 28648823 23537049 27289101 25348373 29652888 28797301 17994087 25830018 22936249 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 12537569 18057021 24495485 24438588 21292972 18563158
20798317 21347285 20075255 24508170 30249741 21282665 20566863 26823975 25401762 28648823 23537049 27289101 25348373 29652888 28797301 17994087 25830018 22936249 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 12537569 18057021 24495485 24438588 21292972 18563158
EMBL
BABH01000476
KQ459439
KPJ00970.1
KQ460397
KPJ15271.1
ODYU01003599
+ More
SOQ42570.1 AGBW02010629 OWR48148.1 LJIG01009021 KRT83914.1 GEZM01016973 JAV91001.1 KQ971338 EFA01829.1 GEZM01016975 GEZM01016974 GEZM01016972 JAV91002.1 KQ977355 KYN03315.1 KQ979074 KYN22995.1 GL451577 EFN78998.1 ADTU01003242 ADTU01003243 KQ981958 KYN31981.1 KQ976692 KYM77748.1 GL443736 EFN61737.1 KK107168 QOIP01000001 EZA56172.1 RLU26770.1 GEDC01026349 GEDC01018947 JAS10949.1 JAS18351.1 GEBQ01019695 JAT20282.1 KQ414631 KOC67242.1 GBYB01011447 JAG81214.1 GECZ01026070 JAS43699.1 GECU01005482 GECU01003515 JAT02225.1 JAT04192.1 GECL01003037 JAP03087.1 ACPB03002866 ACPB03002867 GFTR01008125 JAW08301.1 GL767538 EFZ13549.1 DS235083 EEB11493.1 KQ435689 KOX81252.1 GDHC01015931 JAQ02698.1 GBHO01024632 JAG18972.1 NNAY01002462 OXU21234.1 KZ288288 PBC29315.1 KB632308 ERL91949.1 CVRI01000001 CRK86591.1 ABLF02028572 ABLF02028580 ABLF02028583 ABLF02028584 GFXV01000113 MBW11918.1 GGMS01016227 MBY85430.1 KQ763581 OAD54935.1 APCN01005549 APCN01005550 APCN01005551 APCN01005552 LRGB01002580 KZS07045.1 GGLE01003780 MBY07906.1 GDIP01155169 JAJ68233.1 JR037029 JR037030 AEY57539.1 AEY57540.1 LBMM01000691 KMQ97745.1 IACF01000567 LAB66333.1 GDIP01150281 JAJ73121.1 GDIQ01209439 JAK42286.1 GDIQ01165115 JAK86610.1 LJIJ01000224 ODN00207.1 GAKP01017692 JAC41260.1 GDIP01113643 JAL90071.1 GDIQ01005234 JAN89503.1 GEGO01003417 JAR91987.1 CP012524 ALC41109.1 GFPF01012772 MAA23918.1 GDIP01088835 JAM14880.1 CH940648 EDW61009.1 JH431861 GBXI01015428 GBXI01015152 JAC98863.1 JAC99139.1 OUUW01000001 SPP74875.1 CM002911 KMY95444.1 KMY95445.1 CM000071 EAL26499.2 KRT03142.1 CH902619 EDV36477.2 CM000362 EDX08015.1 CH954179 EDV55409.1 AE013599 AAM70854.1 CM000158 EDW91297.1 KRJ99775.1 AY052013 X92653 CH963846 EDW72567.1 CH480824 EDW56575.1 UFQS01001151 UFQT01001151 SSX09254.1 SSX29156.1 AXCN02001045 CH933808 EDW09963.1 KRG05011.1 GAMC01016771 GAMC01016770 JAB89784.1 CCAG010012974 JXJN01007633 CH916367 EDW00969.1 HACA01012272 CDW29633.1 ATLV01023902 KE525347 KFB49856.1 GL732588 EFX73796.1 GDHF01022013 GDHF01005618 JAI30301.1 JAI46696.1 GDIP01101580 JAM02135.1 GDHF01009180 GDHF01002154 JAI43134.1 JAI50160.1 JH431494 GGFL01016281 MBW80459.1 GG666489 EEN63811.1 APGK01010396 APGK01010397 APGK01010398 APGK01010399 APGK01010400 APGK01010401 KB738183 ENN82738.1 APGK01022962 APGK01022963 APGK01022964 APGK01022965 APGK01022966 APGK01022967 APGK01024417 APGK01024418 APGK01024419 APGK01024420 APGK01024421 APGK01024422 APGK01030142 APGK01030143 APGK01030144 APGK01030145 APGK01030146 KB740760 KB740545 KB740455 ENN79047.1 ENN80406.1 ENN80625.1 CH479210 EDW32805.1
SOQ42570.1 AGBW02010629 OWR48148.1 LJIG01009021 KRT83914.1 GEZM01016973 JAV91001.1 KQ971338 EFA01829.1 GEZM01016975 GEZM01016974 GEZM01016972 JAV91002.1 KQ977355 KYN03315.1 KQ979074 KYN22995.1 GL451577 EFN78998.1 ADTU01003242 ADTU01003243 KQ981958 KYN31981.1 KQ976692 KYM77748.1 GL443736 EFN61737.1 KK107168 QOIP01000001 EZA56172.1 RLU26770.1 GEDC01026349 GEDC01018947 JAS10949.1 JAS18351.1 GEBQ01019695 JAT20282.1 KQ414631 KOC67242.1 GBYB01011447 JAG81214.1 GECZ01026070 JAS43699.1 GECU01005482 GECU01003515 JAT02225.1 JAT04192.1 GECL01003037 JAP03087.1 ACPB03002866 ACPB03002867 GFTR01008125 JAW08301.1 GL767538 EFZ13549.1 DS235083 EEB11493.1 KQ435689 KOX81252.1 GDHC01015931 JAQ02698.1 GBHO01024632 JAG18972.1 NNAY01002462 OXU21234.1 KZ288288 PBC29315.1 KB632308 ERL91949.1 CVRI01000001 CRK86591.1 ABLF02028572 ABLF02028580 ABLF02028583 ABLF02028584 GFXV01000113 MBW11918.1 GGMS01016227 MBY85430.1 KQ763581 OAD54935.1 APCN01005549 APCN01005550 APCN01005551 APCN01005552 LRGB01002580 KZS07045.1 GGLE01003780 MBY07906.1 GDIP01155169 JAJ68233.1 JR037029 JR037030 AEY57539.1 AEY57540.1 LBMM01000691 KMQ97745.1 IACF01000567 LAB66333.1 GDIP01150281 JAJ73121.1 GDIQ01209439 JAK42286.1 GDIQ01165115 JAK86610.1 LJIJ01000224 ODN00207.1 GAKP01017692 JAC41260.1 GDIP01113643 JAL90071.1 GDIQ01005234 JAN89503.1 GEGO01003417 JAR91987.1 CP012524 ALC41109.1 GFPF01012772 MAA23918.1 GDIP01088835 JAM14880.1 CH940648 EDW61009.1 JH431861 GBXI01015428 GBXI01015152 JAC98863.1 JAC99139.1 OUUW01000001 SPP74875.1 CM002911 KMY95444.1 KMY95445.1 CM000071 EAL26499.2 KRT03142.1 CH902619 EDV36477.2 CM000362 EDX08015.1 CH954179 EDV55409.1 AE013599 AAM70854.1 CM000158 EDW91297.1 KRJ99775.1 AY052013 X92653 CH963846 EDW72567.1 CH480824 EDW56575.1 UFQS01001151 UFQT01001151 SSX09254.1 SSX29156.1 AXCN02001045 CH933808 EDW09963.1 KRG05011.1 GAMC01016771 GAMC01016770 JAB89784.1 CCAG010012974 JXJN01007633 CH916367 EDW00969.1 HACA01012272 CDW29633.1 ATLV01023902 KE525347 KFB49856.1 GL732588 EFX73796.1 GDHF01022013 GDHF01005618 JAI30301.1 JAI46696.1 GDIP01101580 JAM02135.1 GDHF01009180 GDHF01002154 JAI43134.1 JAI50160.1 JH431494 GGFL01016281 MBW80459.1 GG666489 EEN63811.1 APGK01010396 APGK01010397 APGK01010398 APGK01010399 APGK01010400 APGK01010401 KB738183 ENN82738.1 APGK01022962 APGK01022963 APGK01022964 APGK01022965 APGK01022966 APGK01022967 APGK01024417 APGK01024418 APGK01024419 APGK01024420 APGK01024421 APGK01024422 APGK01030142 APGK01030143 APGK01030144 APGK01030145 APGK01030146 KB740760 KB740545 KB740455 ENN79047.1 ENN80406.1 ENN80625.1 CH479210 EDW32805.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000192223
UP000007266
+ More
UP000005203 UP000078542 UP000078492 UP000008237 UP000005205 UP000002358 UP000078541 UP000078540 UP000000311 UP000053097 UP000279307 UP000053825 UP000015103 UP000009046 UP000053105 UP000215335 UP000242457 UP000030742 UP000183832 UP000007819 UP000075840 UP000076858 UP000036403 UP000094527 UP000092553 UP000008792 UP000268350 UP000001819 UP000007801 UP000000304 UP000008711 UP000000803 UP000002282 UP000007798 UP000001292 UP000075886 UP000009192 UP000078200 UP000192221 UP000092445 UP000092444 UP000092443 UP000092460 UP000075880 UP000001070 UP000030765 UP000000305 UP000001554 UP000019118 UP000085678 UP000008744
UP000005203 UP000078542 UP000078492 UP000008237 UP000005205 UP000002358 UP000078541 UP000078540 UP000000311 UP000053097 UP000279307 UP000053825 UP000015103 UP000009046 UP000053105 UP000215335 UP000242457 UP000030742 UP000183832 UP000007819 UP000075840 UP000076858 UP000036403 UP000094527 UP000092553 UP000008792 UP000268350 UP000001819 UP000007801 UP000000304 UP000008711 UP000000803 UP000002282 UP000007798 UP000001292 UP000075886 UP000009192 UP000078200 UP000192221 UP000092445 UP000092444 UP000092443 UP000092460 UP000075880 UP000001070 UP000030765 UP000000305 UP000001554 UP000019118 UP000085678 UP000008744
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9IUU4
A0A194Q7J7
A0A194RH60
A0A2H1VP12
A0A212F329
A0A1W4X7D0
+ More
A0A0T6B9B0 A0A1Y1MZA3 D1ZZM8 A0A087ZXM0 A0A1Y1MZA5 A0A151IJE7 A0A195ECP6 E2BZ92 A0A158NY64 K7IYU1 A0A195EV02 A0A195AZT2 E2AXX3 A0A026WJF4 A0A1B6CC38 A0A1B6L999 A0A0L7R8N4 A0A0C9RE72 A0A1B6F0B4 A0A1B6JSP7 A0A0V0G6Y8 T1HMF2 A0A224XJU4 E9J0P2 E0VDN7 A0A0N1IU94 A0A146L2T0 A0A0A9XE34 A0A232ESA3 A0A2A3EC51 U4UDB0 A0A1J1HF50 J9JZ49 A0A2H8TF53 A0A2S2R6V2 A0A310SKP5 A0A1Y9GLF6 A0A164PQ75 A0A2R5LEI4 A0A0N8AEJ6 V9IB44 A0A0J7NZC1 A0A2P2HX23 A0A0N8AGA7 A0A0P5ISW0 A0A0N8BLU3 A0A1D2N4K5 A0A034VHM9 A0A0P5UL86 A0A0P6IJU7 A0A147BMG7 A0A0M3QUR2 A0A224Z242 A0A0P5WAK1 B4LMR8 T1J5F2 A0A0A1WKU4 A0A3B0J1P0 A0A0J9RJ04 Q28X17 B3MDU2 B4QFM3 B3NJY1 E2QCJ5 B4P9H7 Q7K4B6 B4MKC8 B4I7K0 A0A336KWY0 A0A182QTG0 B4KNB0 W8BJJ3 A0A1A9UVB9 A0A1W4VI06 A0A1B0ACW3 A0A1B0FAZ2 A0A1A9YBE8 A0A1B0B2G1 A0A182IMN2 B4J6R6 A0A0K2TUB6 A0A084WI12 E9H3D7 A0A0K8W6E4 A0A0P5VCQ0 A0A0K8VW81 T1ITR6 A0A2M4DSF7 C3Y7I3 N6TXE2 N6UID8 A0A1S3KH78 B4H4W9
A0A0T6B9B0 A0A1Y1MZA3 D1ZZM8 A0A087ZXM0 A0A1Y1MZA5 A0A151IJE7 A0A195ECP6 E2BZ92 A0A158NY64 K7IYU1 A0A195EV02 A0A195AZT2 E2AXX3 A0A026WJF4 A0A1B6CC38 A0A1B6L999 A0A0L7R8N4 A0A0C9RE72 A0A1B6F0B4 A0A1B6JSP7 A0A0V0G6Y8 T1HMF2 A0A224XJU4 E9J0P2 E0VDN7 A0A0N1IU94 A0A146L2T0 A0A0A9XE34 A0A232ESA3 A0A2A3EC51 U4UDB0 A0A1J1HF50 J9JZ49 A0A2H8TF53 A0A2S2R6V2 A0A310SKP5 A0A1Y9GLF6 A0A164PQ75 A0A2R5LEI4 A0A0N8AEJ6 V9IB44 A0A0J7NZC1 A0A2P2HX23 A0A0N8AGA7 A0A0P5ISW0 A0A0N8BLU3 A0A1D2N4K5 A0A034VHM9 A0A0P5UL86 A0A0P6IJU7 A0A147BMG7 A0A0M3QUR2 A0A224Z242 A0A0P5WAK1 B4LMR8 T1J5F2 A0A0A1WKU4 A0A3B0J1P0 A0A0J9RJ04 Q28X17 B3MDU2 B4QFM3 B3NJY1 E2QCJ5 B4P9H7 Q7K4B6 B4MKC8 B4I7K0 A0A336KWY0 A0A182QTG0 B4KNB0 W8BJJ3 A0A1A9UVB9 A0A1W4VI06 A0A1B0ACW3 A0A1B0FAZ2 A0A1A9YBE8 A0A1B0B2G1 A0A182IMN2 B4J6R6 A0A0K2TUB6 A0A084WI12 E9H3D7 A0A0K8W6E4 A0A0P5VCQ0 A0A0K8VW81 T1ITR6 A0A2M4DSF7 C3Y7I3 N6TXE2 N6UID8 A0A1S3KH78 B4H4W9
PDB
1W3B
E-value=1.10694e-15,
Score=206
Ontologies
GO
Topology
Subcellular location
Membrane
Endoplasmic reticulum
Endoplasmic reticulum
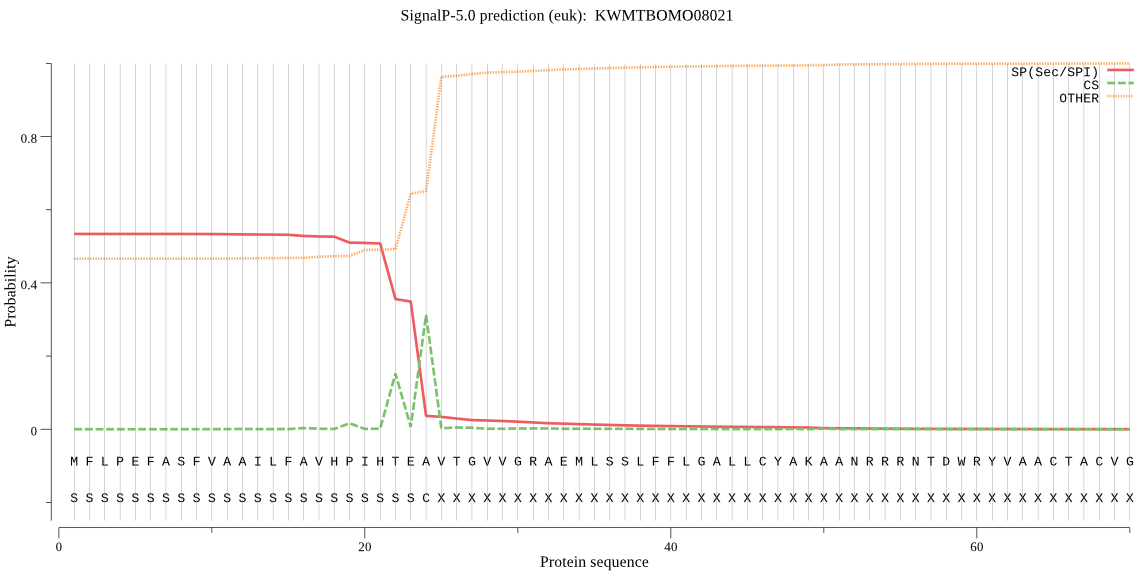
SignalP
Position: 1 - 24,
Likelihood: 0.533350
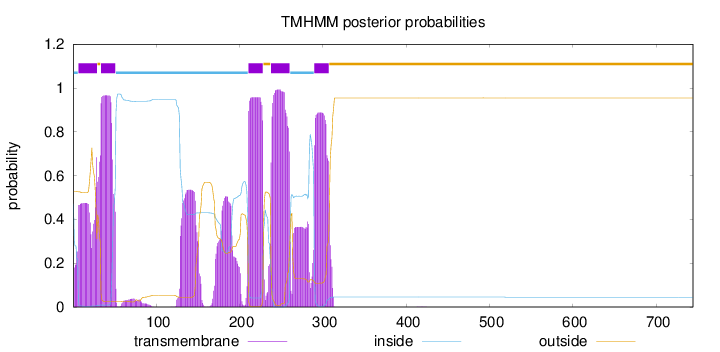
Length:
744
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
117.54205
Exp number, first 60 AAs:
29.52608
Total prob of N-in:
0.47130
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 33
TMhelix
34 - 51
inside
52 - 210
TMhelix
211 - 228
outside
229 - 237
TMhelix
238 - 260
inside
261 - 289
TMhelix
290 - 307
outside
308 - 744
Population Genetic Test Statistics
Pi
24.047772
Theta
21.243985
Tajima's D
0.273501
CLR
0.781551
CSRT
0.459127043647818
Interpretation
Uncertain
