Gene
KWMTBOMO08016 Validated by peptides from experiments
Annotation
cocoonase_precursor_[Bombyx_mori]
Full name
Trypsin 3A1
Location in the cell
Extracellular Reliability : 4.027
Sequence
CDS
ATGGAAAAGTTGTATCTGTTTATTGTTTTTCTTTCATGTGCCTTATTACTGAAAGATGTTACATGTACGGATTCTGAGGCGTTATCCAAAGACGAGGAAAAGATTGTAGGAGGCGAAGAGATTAGCATTAACAAAGTTCCGTACCAAGCGTATCTTTTGCTTCAAAAAGATAACGAATACTTCCAATGCGGAGGTTCGATTATTAGCAAACGTCACATCCTCACGGCGGCACATTGTATCGAAGGTATCTCCAAAGTAACGGTGCGTATCGGAAGCTCAAATTCTAATAAAGGAGGCACCGTTTATACAGCGAAATCAAAGGTGGCTCATCCGAAATACAATTCTAAAACTAAAAACAACGATTTCGCCATTGTCACTGTGAACAAAGACATGGCGATCGATGGAAAAACTACTAAAATCATTACTTTAGCCAAAGAAGGCTCTTCGGTTCCTGACAAAACGAAACTATTGGTTTCCGGGTGGGGAGCTACAAGCGAAGGTGGTTCATCAAGTACAACGCTAAGGGCTGTGCACGTTCAAGCTCATTCTGACGATGAATGCAAGAAATATTTCCGTAGTTTGACATCTAATATGTTCTGCGCTGGGCCCCCCGAAGGCGGAAAGGACTCCTGTCAGGGTGATTCCGGTGGTCCAGCTGTTAAGGGCAATGTCCAACTTGGTGTGGTCTCCTTTGGTGTCGGCTGCGCTCGCAAGAATAACCCTGGTATCTATGCTAAAGTAAGTGCTGCTGCAAAATGGATAAAATCAACGGCGGGCCTATAA
Protein
MEKLYLFIVFLSCALLLKDVTCTDSEALSKDEEKIVGGEEISINKVPYQAYLLLQKDNEYFQCGGSIISKRHILTAAHCIEGISKVTVRIGSSNSNKGGTVYTAKSKVAHPKYNSKTKNNDFAIVTVNKDMAIDGKTTKIITLAKEGSSVPDKTKLLVSGWGATSEGGSSSTTLRAVHVQAHSDDECKKYFRSLTSNMFCAGPPEGGKDSCQGDSGGPAVKGNVQLGVVSFGVGCARKNNPGIYAKVSAAAKWIKSTAGL
Summary
Description
Major function may be to aid in digestion of the blood meal.
Catalytic Activity
Preferential cleavage: Arg-|-Xaa, Lys-|-Xaa.
Similarity
Belongs to the peptidase S1 family.
Keywords
Alternative splicing
Complete proteome
Digestion
Disulfide bond
Hydrolase
Protease
Reference proteome
Secreted
Serine protease
Signal
Zymogen
Feature
propeptide Activation peptide
chain Trypsin 3A1
splice variant In isoform A.
chain Trypsin 3A1
splice variant In isoform A.
Uniprot
A7TVD3
A7UNZ4
E5RVK3
A2TGR7
Q18NR4
D6QUQ4
+ More
A0A3G2LYE2 A0A1B3TP20 A0A1B3TNZ9 A0A1B3TP02 A0A1B3TNW8 A0A1B3TNY5 A0A1B3TP23 A0A3G2LYM8 A0A1B3TNW7 A0A1B3TNY9 A0A1B3TNV7 A0A1B3TP21 A0A194R3X4 A0A3G2LYM4 A0A1B3TNX5 A0A1B3TNW3 A0A1B3TNY2 A0A1B3TNZ8 A0A3S2NEF5 A0A1B3TP00 A0A1B3TNY7 A0A1B3TP08 A0A194Q6G2 A0A1B3TNX6 A0A2A4IV60 A0A1B3TP15 A0A1B3TNZ7 A0A1B3TNV9 A0A1I8Q1B5 A0A1I8MQV4 A0A0J9QZW1 B3N7W6 P29786-2 E3NYI2 G9B5J2 G9B5F5 G9B5E6 G9B5G0 G9B5I6 P29786 G9B5H2 G9B5F7 G9B5F4 G9B5G5 G9B5E8 G9B5H4 G5DGE0 G9B5G4 A0A1L8DQF2 G9B5F2 G9B5J7 Q9VLF5 G9B5G7 G9B5I5 B3DMZ7 A0A1Q3FTA7 U5EZJ4 G9B5F1 B4HYU1 G9B5J8 G9B5E7 A8CW51 G9B5I9 B5AKB2 B8XY09 G9B5H5 G9B5E9 G9B5G8 B4NY05 G9B5I2 G9B5H9 G9B5H8 A0A2H4SAP8 A0A1B0DA37 A0A182GET7 G9B5F6 G9B5G1 G9B5F9 C5IB55 C5IB53 C5IB50 G3JB05 G9B5H1 G9B5J6 C5IB51 C5IB48 C5IB52 G9B5H7 A0A3B3D656 C5IB49 A0A182G8J0 G9B5I7 A0A1I8MB43 Q27J28
A0A3G2LYE2 A0A1B3TP20 A0A1B3TNZ9 A0A1B3TP02 A0A1B3TNW8 A0A1B3TNY5 A0A1B3TP23 A0A3G2LYM8 A0A1B3TNW7 A0A1B3TNY9 A0A1B3TNV7 A0A1B3TP21 A0A194R3X4 A0A3G2LYM4 A0A1B3TNX5 A0A1B3TNW3 A0A1B3TNY2 A0A1B3TNZ8 A0A3S2NEF5 A0A1B3TP00 A0A1B3TNY7 A0A1B3TP08 A0A194Q6G2 A0A1B3TNX6 A0A2A4IV60 A0A1B3TP15 A0A1B3TNZ7 A0A1B3TNV9 A0A1I8Q1B5 A0A1I8MQV4 A0A0J9QZW1 B3N7W6 P29786-2 E3NYI2 G9B5J2 G9B5F5 G9B5E6 G9B5G0 G9B5I6 P29786 G9B5H2 G9B5F7 G9B5F4 G9B5G5 G9B5E8 G9B5H4 G5DGE0 G9B5G4 A0A1L8DQF2 G9B5F2 G9B5J7 Q9VLF5 G9B5G7 G9B5I5 B3DMZ7 A0A1Q3FTA7 U5EZJ4 G9B5F1 B4HYU1 G9B5J8 G9B5E7 A8CW51 G9B5I9 B5AKB2 B8XY09 G9B5H5 G9B5E9 G9B5G8 B4NY05 G9B5I2 G9B5H9 G9B5H8 A0A2H4SAP8 A0A1B0DA37 A0A182GET7 G9B5F6 G9B5G1 G9B5F9 C5IB55 C5IB53 C5IB50 G3JB05 G9B5H1 G9B5J6 C5IB51 C5IB48 C5IB52 G9B5H7 A0A3B3D656 C5IB49 A0A182G8J0 G9B5I7 A0A1I8MB43 Q27J28
EC Number
3.4.21.4
Pubmed
EMBL
EF428980
EU095343
ABR14241.1
ABU49588.1
EU095344
ABU49589.1
+ More
AB604648 BAJ46146.1 EF199627 ABM88498.1 AB257565 BAE96354.1 HM011050 ADG26770.1 MG774893 AYN79048.1 KU925765 AOG75390.1 KU925753 AOG75378.1 KU925749 AOG75374.1 KU925716 AOG75341.1 KU925736 AOG75361.1 KU925769 AOG75394.1 MG774892 AYN79047.1 KU925725 AOG75350.1 KU925739 AOG75364.1 KU925715 AOG75340.1 KU925766 AOG75391.1 KQ460761 KPJ12508.1 MG774891 AYN79046.1 KU925724 AOG75349.1 KU925713 AOG75338.1 KU925737 AOG75362.1 KU925751 AOG75376.1 RSAL01000074 RVE48965.1 KU925750 AOG75375.1 KU925738 AOG75363.1 KU925754 AOG75379.1 KQ459439 KPJ00964.1 KU925726 AOG75351.1 NWSH01005899 PCG63857.1 KU925768 AOG75393.1 KU925752 AOG75377.1 KU925714 AOG75339.1 CM002910 KMY89204.1 CH954177 EDV58327.1 AF487426 AF508783 X64362 CH477469 EF672103 ABW17548.1 HM480752 AEI58598.1 HM480715 AEI58561.1 HM480706 AEI58552.1 HM480720 AEI58566.1 HM480743 HM480746 AEI58592.1 HM480732 AEI58578.1 HM480717 AEI58563.1 HM480714 HM480722 HM480723 HM480726 AEI58560.1 HM480725 AEI58571.1 HM480708 HM480729 AEI58554.1 HM480734 AEI58580.1 JN662341 AEQ55298.1 HM480724 AEI58570.1 GFDF01005376 JAV08708.1 HM480712 AEI58558.1 HM480757 AEI58603.1 AE014134 AAF52738.1 AHN54286.1 HM480727 AEI58573.1 HM480745 AEI58591.1 BT032785 BT032800 ACD81799.1 ACD81814.1 GFDL01004392 JAV30653.1 GANO01000688 JAB59183.1 HM480711 AEI58557.1 CH480818 EDW52221.1 HM480758 AEI58604.1 HM480707 AEI58553.1 AJWK01004381 AJWK01004382 EU124582 ABV60300.1 HM480749 AEI58595.1 EU855138 ACF72874.2 FJ458410 ACL37992.1 HM480735 AEI58581.1 HM480709 AEI58555.1 HM480728 AEI58574.1 CM000157 EDW88607.2 HM480736 HM480742 AEI58588.1 HM480739 AEI58585.1 HM480738 AEI58584.1 CP023323 ATY60163.1 AJVK01028773 JXUM01058532 KQ562007 KXJ76927.1 HM480716 AEI58562.1 HM480721 AEI58567.1 HM480719 AEI58565.1 FJ839170 ACR61226.1 FJ839168 ACR61224.1 FJ839165 ACR61221.1 JH126400 EGX95217.1 HM480731 AEI58577.1 HM480756 AEI58602.1 FJ839166 ACR61222.1 FJ839163 ACR61219.1 FJ839167 ACR61223.1 HM480737 AEI58583.1 FJ839164 FJ839169 FJ839172 ACR61220.1 ACR61225.1 ACR61228.1 JXUM01047815 KQ561534 KXJ78237.1 HM480747 AEI58593.1 DQ396618 ABD57312.1
AB604648 BAJ46146.1 EF199627 ABM88498.1 AB257565 BAE96354.1 HM011050 ADG26770.1 MG774893 AYN79048.1 KU925765 AOG75390.1 KU925753 AOG75378.1 KU925749 AOG75374.1 KU925716 AOG75341.1 KU925736 AOG75361.1 KU925769 AOG75394.1 MG774892 AYN79047.1 KU925725 AOG75350.1 KU925739 AOG75364.1 KU925715 AOG75340.1 KU925766 AOG75391.1 KQ460761 KPJ12508.1 MG774891 AYN79046.1 KU925724 AOG75349.1 KU925713 AOG75338.1 KU925737 AOG75362.1 KU925751 AOG75376.1 RSAL01000074 RVE48965.1 KU925750 AOG75375.1 KU925738 AOG75363.1 KU925754 AOG75379.1 KQ459439 KPJ00964.1 KU925726 AOG75351.1 NWSH01005899 PCG63857.1 KU925768 AOG75393.1 KU925752 AOG75377.1 KU925714 AOG75339.1 CM002910 KMY89204.1 CH954177 EDV58327.1 AF487426 AF508783 X64362 CH477469 EF672103 ABW17548.1 HM480752 AEI58598.1 HM480715 AEI58561.1 HM480706 AEI58552.1 HM480720 AEI58566.1 HM480743 HM480746 AEI58592.1 HM480732 AEI58578.1 HM480717 AEI58563.1 HM480714 HM480722 HM480723 HM480726 AEI58560.1 HM480725 AEI58571.1 HM480708 HM480729 AEI58554.1 HM480734 AEI58580.1 JN662341 AEQ55298.1 HM480724 AEI58570.1 GFDF01005376 JAV08708.1 HM480712 AEI58558.1 HM480757 AEI58603.1 AE014134 AAF52738.1 AHN54286.1 HM480727 AEI58573.1 HM480745 AEI58591.1 BT032785 BT032800 ACD81799.1 ACD81814.1 GFDL01004392 JAV30653.1 GANO01000688 JAB59183.1 HM480711 AEI58557.1 CH480818 EDW52221.1 HM480758 AEI58604.1 HM480707 AEI58553.1 AJWK01004381 AJWK01004382 EU124582 ABV60300.1 HM480749 AEI58595.1 EU855138 ACF72874.2 FJ458410 ACL37992.1 HM480735 AEI58581.1 HM480709 AEI58555.1 HM480728 AEI58574.1 CM000157 EDW88607.2 HM480736 HM480742 AEI58588.1 HM480739 AEI58585.1 HM480738 AEI58584.1 CP023323 ATY60163.1 AJVK01028773 JXUM01058532 KQ562007 KXJ76927.1 HM480716 AEI58562.1 HM480721 AEI58567.1 HM480719 AEI58565.1 FJ839170 ACR61226.1 FJ839168 ACR61224.1 FJ839165 ACR61221.1 JH126400 EGX95217.1 HM480731 AEI58577.1 HM480756 AEI58602.1 FJ839166 ACR61222.1 FJ839163 ACR61219.1 FJ839167 ACR61223.1 HM480737 AEI58583.1 FJ839164 FJ839169 FJ839172 ACR61220.1 ACR61225.1 ACR61228.1 JXUM01047815 KQ561534 KXJ78237.1 HM480747 AEI58593.1 DQ396618 ABD57312.1
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
A7TVD3
A7UNZ4
E5RVK3
A2TGR7
Q18NR4
D6QUQ4
+ More
A0A3G2LYE2 A0A1B3TP20 A0A1B3TNZ9 A0A1B3TP02 A0A1B3TNW8 A0A1B3TNY5 A0A1B3TP23 A0A3G2LYM8 A0A1B3TNW7 A0A1B3TNY9 A0A1B3TNV7 A0A1B3TP21 A0A194R3X4 A0A3G2LYM4 A0A1B3TNX5 A0A1B3TNW3 A0A1B3TNY2 A0A1B3TNZ8 A0A3S2NEF5 A0A1B3TP00 A0A1B3TNY7 A0A1B3TP08 A0A194Q6G2 A0A1B3TNX6 A0A2A4IV60 A0A1B3TP15 A0A1B3TNZ7 A0A1B3TNV9 A0A1I8Q1B5 A0A1I8MQV4 A0A0J9QZW1 B3N7W6 P29786-2 E3NYI2 G9B5J2 G9B5F5 G9B5E6 G9B5G0 G9B5I6 P29786 G9B5H2 G9B5F7 G9B5F4 G9B5G5 G9B5E8 G9B5H4 G5DGE0 G9B5G4 A0A1L8DQF2 G9B5F2 G9B5J7 Q9VLF5 G9B5G7 G9B5I5 B3DMZ7 A0A1Q3FTA7 U5EZJ4 G9B5F1 B4HYU1 G9B5J8 G9B5E7 A8CW51 G9B5I9 B5AKB2 B8XY09 G9B5H5 G9B5E9 G9B5G8 B4NY05 G9B5I2 G9B5H9 G9B5H8 A0A2H4SAP8 A0A1B0DA37 A0A182GET7 G9B5F6 G9B5G1 G9B5F9 C5IB55 C5IB53 C5IB50 G3JB05 G9B5H1 G9B5J6 C5IB51 C5IB48 C5IB52 G9B5H7 A0A3B3D656 C5IB49 A0A182G8J0 G9B5I7 A0A1I8MB43 Q27J28
A0A3G2LYE2 A0A1B3TP20 A0A1B3TNZ9 A0A1B3TP02 A0A1B3TNW8 A0A1B3TNY5 A0A1B3TP23 A0A3G2LYM8 A0A1B3TNW7 A0A1B3TNY9 A0A1B3TNV7 A0A1B3TP21 A0A194R3X4 A0A3G2LYM4 A0A1B3TNX5 A0A1B3TNW3 A0A1B3TNY2 A0A1B3TNZ8 A0A3S2NEF5 A0A1B3TP00 A0A1B3TNY7 A0A1B3TP08 A0A194Q6G2 A0A1B3TNX6 A0A2A4IV60 A0A1B3TP15 A0A1B3TNZ7 A0A1B3TNV9 A0A1I8Q1B5 A0A1I8MQV4 A0A0J9QZW1 B3N7W6 P29786-2 E3NYI2 G9B5J2 G9B5F5 G9B5E6 G9B5G0 G9B5I6 P29786 G9B5H2 G9B5F7 G9B5F4 G9B5G5 G9B5E8 G9B5H4 G5DGE0 G9B5G4 A0A1L8DQF2 G9B5F2 G9B5J7 Q9VLF5 G9B5G7 G9B5I5 B3DMZ7 A0A1Q3FTA7 U5EZJ4 G9B5F1 B4HYU1 G9B5J8 G9B5E7 A8CW51 G9B5I9 B5AKB2 B8XY09 G9B5H5 G9B5E9 G9B5G8 B4NY05 G9B5I2 G9B5H9 G9B5H8 A0A2H4SAP8 A0A1B0DA37 A0A182GET7 G9B5F6 G9B5G1 G9B5F9 C5IB55 C5IB53 C5IB50 G3JB05 G9B5H1 G9B5J6 C5IB51 C5IB48 C5IB52 G9B5H7 A0A3B3D656 C5IB49 A0A182G8J0 G9B5I7 A0A1I8MB43 Q27J28
PDB
2BYA
E-value=2.71279e-34,
Score=361
Ontologies
GO
Topology
Subcellular location
SignalP
Position: 1 - 22,
Likelihood: 0.996385
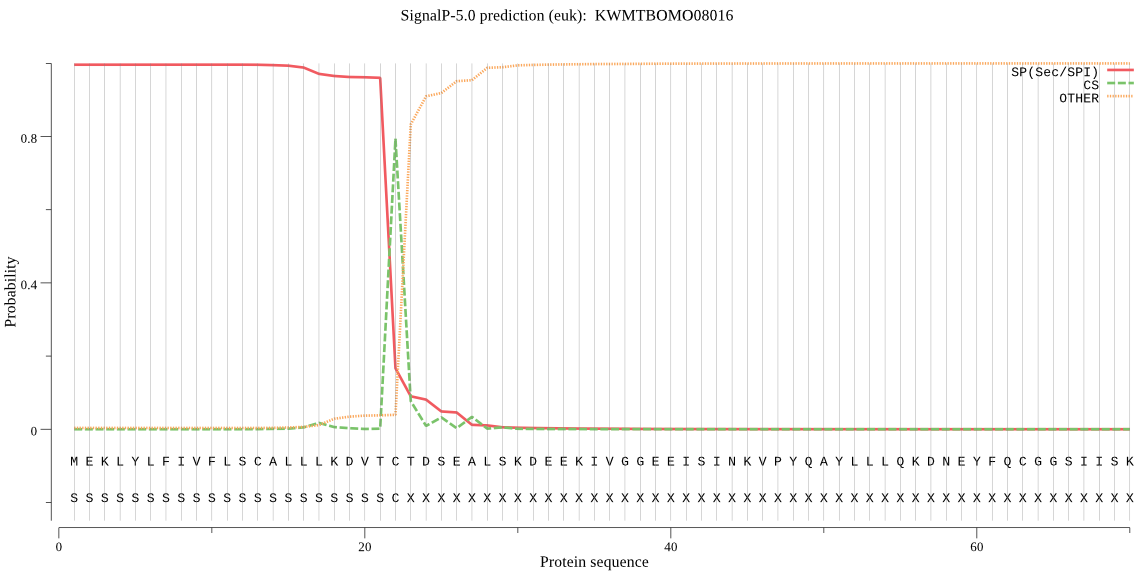
Length:
260
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02456
Exp number, first 60 AAs:
0.02382
Total prob of N-in:
0.00842
outside
1 - 260
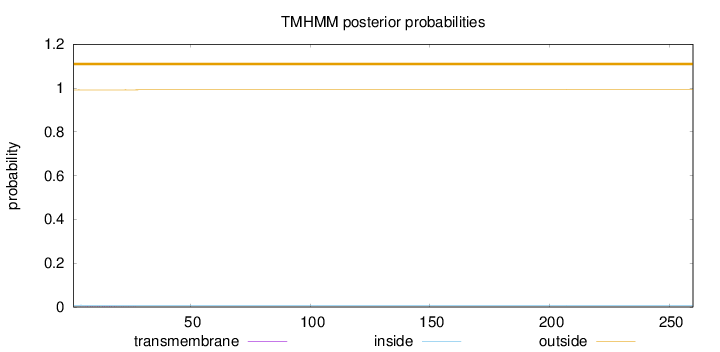
Population Genetic Test Statistics
Pi
20.445042
Theta
23.662266
Tajima's D
-0.521817
CLR
0.315301
CSRT
0.23908804559772
Interpretation
Uncertain
