Gene
KWMTBOMO08015
Annotation
PREDICTED:_uncharacterized_protein_LOC105841315_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.25 Nuclear Reliability : 1.793
Sequence
CDS
ATGAATCCACTTGAATCTTCACTGGCTGAGATGTCAGAGCTGTTTAATGCACGTATGGCGAAGTTCGAAGCCCAGCTCAACAAGTCTCCAGAACTATCAACCACATCATCAAGTTTGGCATTGGAATTTTCGTCATTTCGGGCATTCATTACTGAGGCACTTAATAACTTGCAACAGCAGGTGGAGATGCTCGCGCGCATGCAGGATAATACAGAGACTCGCAGCCGTAGAAAAATGTTGATCATTCACGGAGTAGCTGAGACTAAGGGTGAGCAAATTGAACAGGTATTGTCTAATTTCATAGGCAGTCATCTTGCTTTGGCCGAGTTCTCCTCAGCTGATATTAGAAGATGTCACCGTATGAATAGATTGTCAACATCCGAAGGCTCTCGTCCCATACTGATAAAATTCCAAACGTATGAAATGCGCAGTCAAGTTTGGTTTTCTAAAACAAAACTAAAGGGTACCGGCTTTACTATTTCGGAGTTTCTGACGAAAGCGCGGCACGACATTTTCATGGCTGCACGGAGTAGATTCGGCATGAAGAAGGTCTGGACCAAGGATGGCTGTGTCTACATTTTGGGAGCGGATGGCATCAAACATCGCGTCGTTACACATCGAGATCTTAATCATCTTCAAAAGAACACATCGGAGGCAGTCCCTGAGGTTGATGCTCATAAGCGTGCTGTTACAAAACCGGCTGTGGTAAAGGCAAAGCGCGCTGCTGCTAGAAAGTGA
Protein
MNPLESSLAEMSELFNARMAKFEAQLNKSPELSTTSSSLALEFSSFRAFITEALNNLQQQVEMLARMQDNTETRSRRKMLIIHGVAETKGEQIEQVLSNFIGSHLALAEFSSADIRRCHRMNRLSTSEGSRPILIKFQTYEMRSQVWFSKTKLKGTGFTISEFLTKARHDIFMAARSRFGMKKVWTKDGCVYILGADGIKHRVVTHRDLNHLQKNTSEAVPEVDAHKRAVTKPAVVKAKRAAARK
Summary
Uniprot
A0A2A4J0P1
A0A2A4JPZ6
A0A2A4JP60
A0A2W1AYL0
A0A2A4JF70
A0A2W1C1G2
+ More
A0A2H1W7N3 A0A0L7KPM5 A0A0L7L318 A0A0L7KIU7 A0A0L7K490 A0A2H1X0E0 H9JND1 A0A0L7KW56 A0A0L7K245 A0A0L7LEM3 A0A0L7KMJ9 A0A2H1VLT4 A0A0L7KXD0 A0A0L7K1Y0 A0A0L7L7Q1 A0A0L7LF72 S4PY07 A0A2H1W136 A0A0L7KXE0 A0A2W1B638 A0A2A4JRK8 A0A2A4JAE4 A0A0L7KZD4 A0A2W1BHY6 A0A0L7KWV6 A0A0L7LRK8 A0A2A4JKL5 A0A2A4JSS2 A0A2A4J7K0 A0A2A4JNA4 A0A2A4JVB8 A0A2W1BPW3 A0A2A4JVW6 A0A2A4K0M9 A0A2A4JIA9 A0A2A4ISJ1 A0A2W1BKL8 A0A3S2N3S1 A0A0L7LN09 A0A0L7LT08 A0A1S3DR71 A0A2A4IZX0
A0A2H1W7N3 A0A0L7KPM5 A0A0L7L318 A0A0L7KIU7 A0A0L7K490 A0A2H1X0E0 H9JND1 A0A0L7KW56 A0A0L7K245 A0A0L7LEM3 A0A0L7KMJ9 A0A2H1VLT4 A0A0L7KXD0 A0A0L7K1Y0 A0A0L7L7Q1 A0A0L7LF72 S4PY07 A0A2H1W136 A0A0L7KXE0 A0A2W1B638 A0A2A4JRK8 A0A2A4JAE4 A0A0L7KZD4 A0A2W1BHY6 A0A0L7KWV6 A0A0L7LRK8 A0A2A4JKL5 A0A2A4JSS2 A0A2A4J7K0 A0A2A4JNA4 A0A2A4JVB8 A0A2W1BPW3 A0A2A4JVW6 A0A2A4K0M9 A0A2A4JIA9 A0A2A4ISJ1 A0A2W1BKL8 A0A3S2N3S1 A0A0L7LN09 A0A0L7LT08 A0A1S3DR71 A0A2A4IZX0
EMBL
NWSH01004248
PCG65306.1
NWSH01000800
PCG74117.1
NWSH01000964
PCG73374.1
+ More
KZ150737 PZC70349.1 NWSH01001639 PCG70611.1 KZ149916 PZC77943.1 ODYU01006869 SOQ49099.1 JTDY01007405 KOB65227.1 JTDY01003374 KOB69654.1 JTDY01009601 KOB63242.1 JTDY01010801 JTDY01009435 KOB57999.1 KOB63512.1 ODYU01012456 SOQ58773.1 BABH01019446 BABH01019447 BABH01019448 BABH01019449 BABH01019450 JTDY01005020 KOB67468.1 JTDY01015832 KOB51930.1 JTDY01001444 KOB73845.1 JTDY01009052 KOB64154.1 ODYU01003266 SOQ41795.1 JTDY01004770 KOB67785.1 JTDY01016995 KOB51870.1 JTDY01002460 KOB71369.1 KOB73846.1 GAIX01004118 JAA88442.1 ODYU01005692 SOQ46801.1 JTDY01004617 KOB67938.1 KZ150731 PZC70355.1 NWSH01000709 PCG74665.1 NWSH01002122 PCG69097.1 JTDY01004162 KOB68510.1 KZ150113 PZC73314.1 JTDY01004815 JTDY01001265 KOB67717.1 KOB74342.1 JTDY01000235 KOB78128.1 NWSH01001172 PCG72268.1 NWSH01000695 PCG74738.1 NWSH01002863 PCG67420.1 NWSH01000981 PCG73266.1 NWSH01000574 PCG75564.1 KZ149998 PZC75357.1 NWSH01000512 PCG75926.1 NWSH01000281 PCG77797.1 NWSH01001465 PCG71162.1 PCG71163.1 NWSH01008089 PCG62701.1 KZ150083 PZC73827.1 RSAL01001827 RVE40669.1 JTDY01000521 KOB76815.1 JTDY01000147 KOB78598.1 NWSH01004711 PCG64730.1
KZ150737 PZC70349.1 NWSH01001639 PCG70611.1 KZ149916 PZC77943.1 ODYU01006869 SOQ49099.1 JTDY01007405 KOB65227.1 JTDY01003374 KOB69654.1 JTDY01009601 KOB63242.1 JTDY01010801 JTDY01009435 KOB57999.1 KOB63512.1 ODYU01012456 SOQ58773.1 BABH01019446 BABH01019447 BABH01019448 BABH01019449 BABH01019450 JTDY01005020 KOB67468.1 JTDY01015832 KOB51930.1 JTDY01001444 KOB73845.1 JTDY01009052 KOB64154.1 ODYU01003266 SOQ41795.1 JTDY01004770 KOB67785.1 JTDY01016995 KOB51870.1 JTDY01002460 KOB71369.1 KOB73846.1 GAIX01004118 JAA88442.1 ODYU01005692 SOQ46801.1 JTDY01004617 KOB67938.1 KZ150731 PZC70355.1 NWSH01000709 PCG74665.1 NWSH01002122 PCG69097.1 JTDY01004162 KOB68510.1 KZ150113 PZC73314.1 JTDY01004815 JTDY01001265 KOB67717.1 KOB74342.1 JTDY01000235 KOB78128.1 NWSH01001172 PCG72268.1 NWSH01000695 PCG74738.1 NWSH01002863 PCG67420.1 NWSH01000981 PCG73266.1 NWSH01000574 PCG75564.1 KZ149998 PZC75357.1 NWSH01000512 PCG75926.1 NWSH01000281 PCG77797.1 NWSH01001465 PCG71162.1 PCG71163.1 NWSH01008089 PCG62701.1 KZ150083 PZC73827.1 RSAL01001827 RVE40669.1 JTDY01000521 KOB76815.1 JTDY01000147 KOB78598.1 NWSH01004711 PCG64730.1
Proteomes
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4J0P1
A0A2A4JPZ6
A0A2A4JP60
A0A2W1AYL0
A0A2A4JF70
A0A2W1C1G2
+ More
A0A2H1W7N3 A0A0L7KPM5 A0A0L7L318 A0A0L7KIU7 A0A0L7K490 A0A2H1X0E0 H9JND1 A0A0L7KW56 A0A0L7K245 A0A0L7LEM3 A0A0L7KMJ9 A0A2H1VLT4 A0A0L7KXD0 A0A0L7K1Y0 A0A0L7L7Q1 A0A0L7LF72 S4PY07 A0A2H1W136 A0A0L7KXE0 A0A2W1B638 A0A2A4JRK8 A0A2A4JAE4 A0A0L7KZD4 A0A2W1BHY6 A0A0L7KWV6 A0A0L7LRK8 A0A2A4JKL5 A0A2A4JSS2 A0A2A4J7K0 A0A2A4JNA4 A0A2A4JVB8 A0A2W1BPW3 A0A2A4JVW6 A0A2A4K0M9 A0A2A4JIA9 A0A2A4ISJ1 A0A2W1BKL8 A0A3S2N3S1 A0A0L7LN09 A0A0L7LT08 A0A1S3DR71 A0A2A4IZX0
A0A2H1W7N3 A0A0L7KPM5 A0A0L7L318 A0A0L7KIU7 A0A0L7K490 A0A2H1X0E0 H9JND1 A0A0L7KW56 A0A0L7K245 A0A0L7LEM3 A0A0L7KMJ9 A0A2H1VLT4 A0A0L7KXD0 A0A0L7K1Y0 A0A0L7L7Q1 A0A0L7LF72 S4PY07 A0A2H1W136 A0A0L7KXE0 A0A2W1B638 A0A2A4JRK8 A0A2A4JAE4 A0A0L7KZD4 A0A2W1BHY6 A0A0L7KWV6 A0A0L7LRK8 A0A2A4JKL5 A0A2A4JSS2 A0A2A4J7K0 A0A2A4JNA4 A0A2A4JVB8 A0A2W1BPW3 A0A2A4JVW6 A0A2A4K0M9 A0A2A4JIA9 A0A2A4ISJ1 A0A2W1BKL8 A0A3S2N3S1 A0A0L7LN09 A0A0L7LT08 A0A1S3DR71 A0A2A4IZX0
Ontologies
KEGG
PANTHER
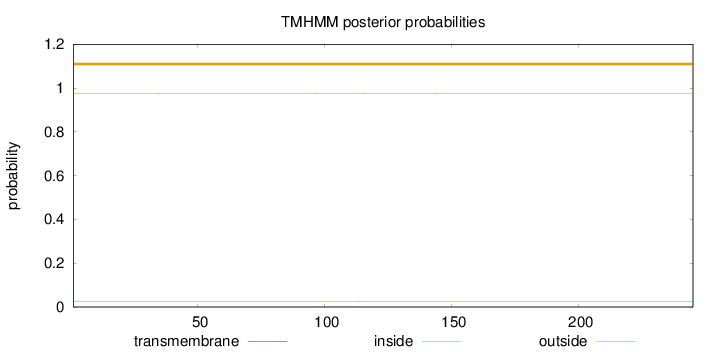
Topology
Length:
245
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00690999999999999
Exp number, first 60 AAs:
0.00183
Total prob of N-in:
0.02409
outside
1 - 245
Population Genetic Test Statistics
Pi
222.705912
Theta
146.141454
Tajima's D
1.749062
CLR
0
CSRT
0.84040797960102
Interpretation
Uncertain
