Gene
KWMTBOMO08013
Pre Gene Modal
BGIBMGA001135
Annotation
PREDICTED:_uncharacterized_protein_LOC101741917_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.358
Sequence
CDS
ATGGATCGTCGACTAATTGCTCTTTTGGCGGTGGCCGCCCATTTAAGTTGTATGACACCAGTACGAGCACAAGGAAATCTCCTCACTCAGACTGTCTATGGATTCTTGGACTTCACGACAACCATTGGCAATACCGTAATGGTGTTCTCGCCCCAGTCAGCGCCTCCTCCAGAACCGCCCAAAACTGAAAAGTCAGTTCAAGAAAATATCATCGAAACGAAACCTCCACCGCCATCAGTTACAAATGTCAACCCGGAAGCCATCAAACCTAGTAAACTGTCAGACAAAAACAAAACCAATAAGCCGGCTCCACCAGCTGTTGTGTCAAGTGCAGTAAACGTGGTCTCTTCATCTCCTAAAAAAGAGGAAAAGAAAGGAAACACTCCAAAGTCAACAATTAAAGAACAAAGCCAAGTCATCACAAAAGTCGAAGTAGTAGCAGGACCTTCAAAAATAGTAGAAAGTAGTTCACCAAAACAAAAATCAAATAAACAGAACTCTCAATCTAGTAACAAGAATAAAAAGAACGAAATTAAGAGCGTTACAAACATCGTGAGCAGTAAAGTTGAAGTAAAACAAGGCACGCCACCGTCGATAATAAGCTCCAAAATCAATAGCGTTGTTCAAGTTAAATCGAGTCAGTCTGACGAAGAACCAGCTATATTAATCTCGAACAATAATTTAGGCGAACCGGAATACGATTTCTTGTCGCGACAACCCTCTGAATTTGTTGAAGAAACATACAAAGTAATTAACATCAGACCCTCTAAAACTCATGGATCGAAGCCAAAGCACGCTCATAAGAATAGAGCCCACCAGCCTAGCCCTCCTTCGGATGATGACGAACACCCTCTCGGCCTTGTAACTACTTTAGGCGGTACCATCGTCAAAGATGGACTTACAACGATCCATGAAACAAGTGTCATAGGAACATACATTTCTGGAAAATATGCACAAGTTTTAAACAGTAACTCTAAAATTATACAACCGGCTGGTCATAGAGCCAAAATATCTCCCTCTCCCACACACAGAATTCTTAAAACAGCCGCACCGGCTATTTCTAAAAATCACAGACACAACTTGGAACCAACTCCCTCTATTTCTGATGACGATAGTAGCTTTGGTAAAACCTCACGCAGACAAACAATAAGTGGCAGCTCATTTAAAAATCGACACCGATCCCAAAACTTTGCTGAAAACGATAATCACGACAACAGTCCTGCTAATCAAAGCAAAAAATTCAAAAACCGAAATGCCCAGAGGACTCCCAGTGAAACAGCGACACCGTCATTCTCTAATAGAAGAAAGAGCAGTCGAAATTCAGGTCACAAATCCGTGGTGACTCCCGCCAACAACAACGAGAATCAGTCTCGGCGAGGATTCAAACCTCGAGCTCAGCCATCTCCAGTCGAACAAGTGACTCCAGCACCGTCAACAAGTCTTTATAAATTTAAATTGAACCGAACTCCGGGTTCGGGACGATGGCAGTACAAAACTAGTCCTAAACCGAAAGTAACCATCCGTAAGGCCGTAAGTGAGGAGGAGCTGCAACAAACGACCCCTAATACCAACCCTCTTCTGGATGATACGCAATTAAACGATATTTCTCCTCAAGCAAGATCAGATAACGATCTTGAGCTCTCTGGATCTCAAAGTGGTCCAGGCACGCTACTTGACAATGACACTGAGGACAATTCAATTGAAAAACTGCCACCAGTTGAAACACTAAAAGTCGAAATTTCCACACCTGCCGACTTCAGTGACGTGTATTACGAAATTGCAACAATTAAATCGCCCTATACTTTCCAGGTTGGACGTGTTAAAAATACTCGTGTTATAACAGTAACCTCAACAATAGAAAAACGTTTAGAACCAACAGTAGCTCCATCGCAAATTTCTCTCAACGAGCCGTTGACAGAAAACATATTGGCGACCGCATCTCCCTATGGCAAAGATCACAATTTAGATTCAAGCATCGCTACTCTACCAGCTATAACGCTGCCTTCTGACATGGAAACCCCGCCACTTGAAACTATTACTGAAACATTCAGTACAACACAGAACATGCTTAAAACTCACATTCTGCCCGTAGTAAGAGACGTCAACATGACAAGTAGCTTGACTTTTATTCAAACATACCAAATCACAAGATTCGTTACGGCAACGAAGACTTTACCACCAATGGACTACTTCCAATTTATACCCAGCAAAACATTAAAGGAATTCAACAGCCGACTGGACGAAGCAGGTTCAGAATTACATTTGGAACTAGATTTTGGAGATAGCACCGAAGACGAAGACGGTGTTCCTCGACGAGTCTTCCCACCAGAATTGGATCTCGCTAATATTGGCTCTGATTTTGATTTAACCGAAATCGACAAATACGGAGCAGACAACCATTTAAGGCTCAAAAAAGCACACGGACAAAACAAAGCAAACCATGTAACGGAAGCGCCAAACGTGCCGACTCCTGCTTTAACTCCCGAACAAGCGCAACAATTGGCTCTGCTTAGACTCCTCAATCCAGCCGCTGCAGCTCAAATACCTAACGTCGTTACGACTTCGAAACCTATTTTCAAATACGAAACTATTTATGAATCGCATGTCATTCCATTCTTCGACGGAAGAAATACAGTGCAAAGCACCATTTCACGACCCATTGCAACTGTCACGAAAACAGAGTACGAAATAGGAACGAGTAGTTTGCCTGCTTTGCCTCTGCAGCCGATCAATCCTTTGTTCCCTCAACAACAGTTTACTCTAATTTCGACACCGGTCGTCATGAACACCGAAGTAATAGCAACTGATAGTCAAATTCTAAAATTAACTTTCGGTGCGAAAACCGTGTACACGACATTGTTCACGACCAAAGTCGTACCGACCGTGCTTACTTCCTACGTAACCTCATCGATACCTGTACAAGCCTCGGCGGGCTACCCGGGATATTTCCCGCCACCCTTCGCACCTTACCCATATGTAGGTTAA
Protein
MDRRLIALLAVAAHLSCMTPVRAQGNLLTQTVYGFLDFTTTIGNTVMVFSPQSAPPPEPPKTEKSVQENIIETKPPPPSVTNVNPEAIKPSKLSDKNKTNKPAPPAVVSSAVNVVSSSPKKEEKKGNTPKSTIKEQSQVITKVEVVAGPSKIVESSSPKQKSNKQNSQSSNKNKKNEIKSVTNIVSSKVEVKQGTPPSIISSKINSVVQVKSSQSDEEPAILISNNNLGEPEYDFLSRQPSEFVEETYKVINIRPSKTHGSKPKHAHKNRAHQPSPPSDDDEHPLGLVTTLGGTIVKDGLTTIHETSVIGTYISGKYAQVLNSNSKIIQPAGHRAKISPSPTHRILKTAAPAISKNHRHNLEPTPSISDDDSSFGKTSRRQTISGSSFKNRHRSQNFAENDNHDNSPANQSKKFKNRNAQRTPSETATPSFSNRRKSSRNSGHKSVVTPANNNENQSRRGFKPRAQPSPVEQVTPAPSTSLYKFKLNRTPGSGRWQYKTSPKPKVTIRKAVSEEELQQTTPNTNPLLDDTQLNDISPQARSDNDLELSGSQSGPGTLLDNDTEDNSIEKLPPVETLKVEISTPADFSDVYYEIATIKSPYTFQVGRVKNTRVITVTSTIEKRLEPTVAPSQISLNEPLTENILATASPYGKDHNLDSSIATLPAITLPSDMETPPLETITETFSTTQNMLKTHILPVVRDVNMTSSLTFIQTYQITRFVTATKTLPPMDYFQFIPSKTLKEFNSRLDEAGSELHLELDFGDSTEDEDGVPRRVFPPELDLANIGSDFDLTEIDKYGADNHLRLKKAHGQNKANHVTEAPNVPTPALTPEQAQQLALLRLLNPAAAAQIPNVVTTSKPIFKYETIYESHVIPFFDGRNTVQSTISRPIATVTKTEYEIGTSSLPALPLQPINPLFPQQQFTLISTPVVMNTEVIATDSQILKLTFGAKTVYTTLFTTKVVPTVLTSYVTSSIPVQASAGYPGYFPPPFAPYPYVG
Summary
Uniprot
H9IV55
A0A2A4J2J2
A0A3S2LL08
A0A2H1V5C1
A0A194R5J5
A0A194Q5Z9
+ More
A0A212FP35 A0A0L7KY58 A0A0T6B9B8 D1ZZN4 A0A1Y1M473 A0A1J1IZJ4 A0A336LVS5 A0A182JUW9 A0A182FV18 A0A182MYF7 A0A182P8F2 N6UNB2 N6UU90 A0A182V1Y5 A0A182XCE0 A0A182U2C8 A0A182HFP3 A0A182LQ51 A0A182RKZ2 A0A182WCW3 A0A182SG21 A0A084WI28 A0A182YA55 A0A182Q0M9 W5JSP1 A0A182LV10 A0A0J7L6Q0 A0A158NER1 A0A195E030 A0A195CRQ1 F4WRN5 A0A195AVR8 A0A195FMZ2 Q17MP5 E2A156 E9IAA0 E2B774 A0A151XIK4 K7J2U0 A0A154PN07 A0A088A8Z1 A0A026WDF7 A0A0C9PZ19 A0A0M8ZXA1 Q5TSF6 A0A0L7QY83 J9JKF9 A0A2S2QVT4 A0A2A3E4E2 A0A2R7VUV3 B4MQQ5 T1HN18 B4J6D8 A0A1W4V1A0 A0A0J9RK67 B4I2D2 B3NQ78 Q7JRF0 A0A0Q5VNR1 B4PAU1 A0A0Q9WLD4 A0A0Q9WXI3 B4KMA7 Q28ZY9
A0A212FP35 A0A0L7KY58 A0A0T6B9B8 D1ZZN4 A0A1Y1M473 A0A1J1IZJ4 A0A336LVS5 A0A182JUW9 A0A182FV18 A0A182MYF7 A0A182P8F2 N6UNB2 N6UU90 A0A182V1Y5 A0A182XCE0 A0A182U2C8 A0A182HFP3 A0A182LQ51 A0A182RKZ2 A0A182WCW3 A0A182SG21 A0A084WI28 A0A182YA55 A0A182Q0M9 W5JSP1 A0A182LV10 A0A0J7L6Q0 A0A158NER1 A0A195E030 A0A195CRQ1 F4WRN5 A0A195AVR8 A0A195FMZ2 Q17MP5 E2A156 E9IAA0 E2B774 A0A151XIK4 K7J2U0 A0A154PN07 A0A088A8Z1 A0A026WDF7 A0A0C9PZ19 A0A0M8ZXA1 Q5TSF6 A0A0L7QY83 J9JKF9 A0A2S2QVT4 A0A2A3E4E2 A0A2R7VUV3 B4MQQ5 T1HN18 B4J6D8 A0A1W4V1A0 A0A0J9RK67 B4I2D2 B3NQ78 Q7JRF0 A0A0Q5VNR1 B4PAU1 A0A0Q9WLD4 A0A0Q9WXI3 B4KMA7 Q28ZY9
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
28004739 23537049 20966253 24438588 25244985 20920257 23761445 21347285 21719571 17510324 20798317 21282665 20075255 24508170 30249741 12364791 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 15632085 23185243
28004739 23537049 20966253 24438588 25244985 20920257 23761445 21347285 21719571 17510324 20798317 21282665 20075255 24508170 30249741 12364791 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 15632085 23185243
EMBL
BABH01000544
NWSH01003977
PCG65603.1
RSAL01000074
RVE48963.1
ODYU01000577
+ More
SOQ35582.1 KQ460761 KPJ12510.1 KQ459439 KPJ00962.1 AGBW02004200 OWR55469.1 JTDY01004585 KOB67984.1 LJIG01009021 KRT83913.1 KQ971338 EFA01825.2 GEZM01042462 JAV79838.1 CVRI01000064 CRL04972.1 UFQT01000227 SSX22064.1 APGK01015567 APGK01015568 APGK01015569 APGK01015570 APGK01015571 KB739617 ENN82196.1 APGK01013830 APGK01013831 APGK01013832 APGK01013833 APGK01013834 KB739123 ENN82372.1 APCN01005560 APCN01005561 APCN01005562 ATLV01023909 KE525347 KFB49872.1 AXCN02001046 ADMH02000480 ETN66323.1 AXCM01003445 AXCM01003446 LBMM01000509 KMQ98184.1 ADTU01013611 KQ979999 KYN18307.1 KQ977381 KYN03172.1 GL888292 EGI63042.1 KQ976736 KYM76120.1 KQ981490 KYN41279.1 CH477204 EAT47977.1 GL435707 EFN72823.1 GL761997 EFZ22500.1 GL446149 EFN88435.1 KQ982080 KYQ60243.1 KQ434996 KZC13232.1 KK107260 QOIP01000001 EZA54003.1 RLU27363.1 GBYB01006753 JAG76520.1 KQ435827 KOX71946.1 AAAB01008933 EAL40359.3 KQ414693 KOC63568.1 ABLF02014897 ABLF02020278 ABLF02032583 ABLF02032584 ABLF02032586 ABLF02032587 ABLF02032589 ABLF02032591 ABLF02046780 ABLF02061610 GGMS01012631 MBY81834.1 KZ288379 PBC26558.1 KK854100 PTY11274.1 CH963849 EDW74444.1 ACPB03002863 CH916367 EDW01939.1 CM002911 KMY96256.1 CH480820 EDW53927.1 CH954179 EDV56951.1 AE013599 BT001664 AAF47161.1 AAN71419.1 KQS63101.1 CM000158 EDW92481.1 KRK00544.1 CH940659 KRF85676.1 KRF85674.1 KRF85675.1 CH933808 EDW09795.2 KRG04904.1 KRG04905.1 CM000071 EAL25473.3 KRT02246.1 KRT02247.1
SOQ35582.1 KQ460761 KPJ12510.1 KQ459439 KPJ00962.1 AGBW02004200 OWR55469.1 JTDY01004585 KOB67984.1 LJIG01009021 KRT83913.1 KQ971338 EFA01825.2 GEZM01042462 JAV79838.1 CVRI01000064 CRL04972.1 UFQT01000227 SSX22064.1 APGK01015567 APGK01015568 APGK01015569 APGK01015570 APGK01015571 KB739617 ENN82196.1 APGK01013830 APGK01013831 APGK01013832 APGK01013833 APGK01013834 KB739123 ENN82372.1 APCN01005560 APCN01005561 APCN01005562 ATLV01023909 KE525347 KFB49872.1 AXCN02001046 ADMH02000480 ETN66323.1 AXCM01003445 AXCM01003446 LBMM01000509 KMQ98184.1 ADTU01013611 KQ979999 KYN18307.1 KQ977381 KYN03172.1 GL888292 EGI63042.1 KQ976736 KYM76120.1 KQ981490 KYN41279.1 CH477204 EAT47977.1 GL435707 EFN72823.1 GL761997 EFZ22500.1 GL446149 EFN88435.1 KQ982080 KYQ60243.1 KQ434996 KZC13232.1 KK107260 QOIP01000001 EZA54003.1 RLU27363.1 GBYB01006753 JAG76520.1 KQ435827 KOX71946.1 AAAB01008933 EAL40359.3 KQ414693 KOC63568.1 ABLF02014897 ABLF02020278 ABLF02032583 ABLF02032584 ABLF02032586 ABLF02032587 ABLF02032589 ABLF02032591 ABLF02046780 ABLF02061610 GGMS01012631 MBY81834.1 KZ288379 PBC26558.1 KK854100 PTY11274.1 CH963849 EDW74444.1 ACPB03002863 CH916367 EDW01939.1 CM002911 KMY96256.1 CH480820 EDW53927.1 CH954179 EDV56951.1 AE013599 BT001664 AAF47161.1 AAN71419.1 KQS63101.1 CM000158 EDW92481.1 KRK00544.1 CH940659 KRF85676.1 KRF85674.1 KRF85675.1 CH933808 EDW09795.2 KRG04904.1 KRG04905.1 CM000071 EAL25473.3 KRT02246.1 KRT02247.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000007266 UP000183832 UP000075881 UP000069272 UP000075884 UP000075885 UP000019118 UP000075903 UP000076407 UP000075902 UP000075840 UP000075882 UP000075900 UP000075920 UP000075901 UP000030765 UP000076408 UP000075886 UP000000673 UP000075883 UP000036403 UP000005205 UP000078492 UP000078542 UP000007755 UP000078540 UP000078541 UP000008820 UP000000311 UP000008237 UP000075809 UP000002358 UP000076502 UP000005203 UP000053097 UP000279307 UP000053105 UP000007062 UP000053825 UP000007819 UP000242457 UP000007798 UP000015103 UP000001070 UP000192221 UP000001292 UP000008711 UP000000803 UP000002282 UP000008792 UP000009192 UP000001819
UP000037510 UP000007266 UP000183832 UP000075881 UP000069272 UP000075884 UP000075885 UP000019118 UP000075903 UP000076407 UP000075902 UP000075840 UP000075882 UP000075900 UP000075920 UP000075901 UP000030765 UP000076408 UP000075886 UP000000673 UP000075883 UP000036403 UP000005205 UP000078492 UP000078542 UP000007755 UP000078540 UP000078541 UP000008820 UP000000311 UP000008237 UP000075809 UP000002358 UP000076502 UP000005203 UP000053097 UP000279307 UP000053105 UP000007062 UP000053825 UP000007819 UP000242457 UP000007798 UP000015103 UP000001070 UP000192221 UP000001292 UP000008711 UP000000803 UP000002282 UP000008792 UP000009192 UP000001819
Pfam
PF15950 DUF4758
Interpro
IPR031866
DUF4758
ProteinModelPortal
H9IV55
A0A2A4J2J2
A0A3S2LL08
A0A2H1V5C1
A0A194R5J5
A0A194Q5Z9
+ More
A0A212FP35 A0A0L7KY58 A0A0T6B9B8 D1ZZN4 A0A1Y1M473 A0A1J1IZJ4 A0A336LVS5 A0A182JUW9 A0A182FV18 A0A182MYF7 A0A182P8F2 N6UNB2 N6UU90 A0A182V1Y5 A0A182XCE0 A0A182U2C8 A0A182HFP3 A0A182LQ51 A0A182RKZ2 A0A182WCW3 A0A182SG21 A0A084WI28 A0A182YA55 A0A182Q0M9 W5JSP1 A0A182LV10 A0A0J7L6Q0 A0A158NER1 A0A195E030 A0A195CRQ1 F4WRN5 A0A195AVR8 A0A195FMZ2 Q17MP5 E2A156 E9IAA0 E2B774 A0A151XIK4 K7J2U0 A0A154PN07 A0A088A8Z1 A0A026WDF7 A0A0C9PZ19 A0A0M8ZXA1 Q5TSF6 A0A0L7QY83 J9JKF9 A0A2S2QVT4 A0A2A3E4E2 A0A2R7VUV3 B4MQQ5 T1HN18 B4J6D8 A0A1W4V1A0 A0A0J9RK67 B4I2D2 B3NQ78 Q7JRF0 A0A0Q5VNR1 B4PAU1 A0A0Q9WLD4 A0A0Q9WXI3 B4KMA7 Q28ZY9
A0A212FP35 A0A0L7KY58 A0A0T6B9B8 D1ZZN4 A0A1Y1M473 A0A1J1IZJ4 A0A336LVS5 A0A182JUW9 A0A182FV18 A0A182MYF7 A0A182P8F2 N6UNB2 N6UU90 A0A182V1Y5 A0A182XCE0 A0A182U2C8 A0A182HFP3 A0A182LQ51 A0A182RKZ2 A0A182WCW3 A0A182SG21 A0A084WI28 A0A182YA55 A0A182Q0M9 W5JSP1 A0A182LV10 A0A0J7L6Q0 A0A158NER1 A0A195E030 A0A195CRQ1 F4WRN5 A0A195AVR8 A0A195FMZ2 Q17MP5 E2A156 E9IAA0 E2B774 A0A151XIK4 K7J2U0 A0A154PN07 A0A088A8Z1 A0A026WDF7 A0A0C9PZ19 A0A0M8ZXA1 Q5TSF6 A0A0L7QY83 J9JKF9 A0A2S2QVT4 A0A2A3E4E2 A0A2R7VUV3 B4MQQ5 T1HN18 B4J6D8 A0A1W4V1A0 A0A0J9RK67 B4I2D2 B3NQ78 Q7JRF0 A0A0Q5VNR1 B4PAU1 A0A0Q9WLD4 A0A0Q9WXI3 B4KMA7 Q28ZY9
Ontologies
GO
Topology
SignalP
Position: 1 - 23,
Likelihood: 0.974332
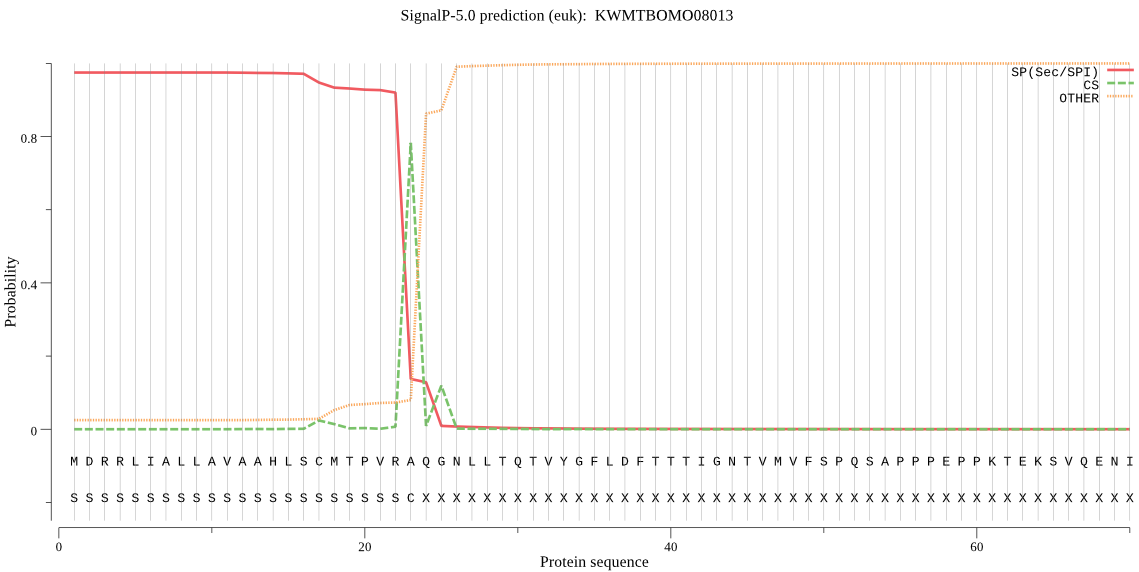
Length:
994
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.90828
Exp number, first 60 AAs:
0.84216
Total prob of N-in:
0.04132
outside
1 - 994

Population Genetic Test Statistics
Pi
300.704882
Theta
215.992353
Tajima's D
1.250463
CLR
0
CSRT
0.728063596820159
Interpretation
Uncertain
