Gene
KWMTBOMO08011 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001136
Annotation
signal_peptidase_complex_subunit_3_[Bombyx_mori]
Full name
Signal peptidase complex subunit 3
Alternative Name
Microsomal signal peptidase 22 kDa subunit
Location in the cell
Extracellular Reliability : 2.295
Sequence
CDS
ATGTATTCCGTGTTAACGAGAGGAAATGCAATACTGACATACACGCTAAGCGTACTAGCATGTCTTACTTTCTTGTGTTTTTTATCAACCCTGACTGTGGACTACAGAACTGGGGCCCAAATGAATACAGTTAAAGTAGTAGTTAAAAACGTACCTGACTACGGAGCATCTAGGGAACGAAATGATCTTGGCTTCTTAACTTTCGATCTTAAAACAGATCTGTCCAACCTCTTTAACTGGAACGTTAAACAGTTGTTCCTATATCTCACTGCCGAATACATTACACCAAGCAATGAATTGAATCAGGTTGTATTGTGGGACAAAATAATATTAAGAGGTGAAAATGCTGTGCTAGACTTCAAGAACATGAACACTAAGTACTATTTCTGGGACGATGGCAACGGCTTAAAAGGTCACAGCAATGTCACATTAACATTATCATGGAATATTATTCCCAATGCTGGCTTGTTACCCAACATCCAGGCTCTTGGTCAACACTCCTTCAAGTTTCCTATAGAATATACTCAAACAAGAGTATGA
Protein
MYSVLTRGNAILTYTLSVLACLTFLCFLSTLTVDYRTGAQMNTVKVVVKNVPDYGASRERNDLGFLTFDLKTDLSNLFNWNVKQLFLYLTAEYITPSNELNQVVLWDKIILRGENAVLDFKNMNTKYYFWDDGNGLKGHSNVTLTLSWNIIPNAGLLPNIQALGQHSFKFPIEYTQTRV
Summary
Description
Component of the microsomal signal peptidase complex which removes signal peptides and other N-terminal peptides from nascent proteins as they are translocated into the lumen of the endoplasmic reticulum.
Subunit
Complex of five different proteins: SEC11L1, SEC11L3, Spase12/SPCS1, Spase25/SPCS2 and Spase22-23/SPCS3.
Similarity
Belongs to the SPCS3 family.
Belongs to the enoyl-CoA hydratase/isomerase family.
Belongs to the enoyl-CoA hydratase/isomerase family.
Keywords
Complete proteome
Endoplasmic reticulum
Glycoprotein
Hydrolase
Membrane
Microsome
Protease
Reference proteome
Signal-anchor
Transmembrane
Transmembrane helix
Feature
chain Signal peptidase complex subunit 3
Uniprot
Q1HQ14
A0A2H1V5P2
A0A2A4J9Q2
A0A1E1WS34
I4DK09
A0A3S2M1P9
+ More
A0A212FP10 S4PYE5 E0VVU3 A0A1B6K719 A0A1B6FD55 U5EZA3 A0A1B6DLS3 T1E333 B4NAG0 A0A1B6KPJ9 A0A1B0EY93 A0A1L8DV91 Q1HRQ4 B4QT28 A0A182Q241 B4HGK6 A0A1B0CYQ9 A0A1S4HE52 A0A2C9GRH5 A0A182USP7 A0A182U3B1 A0A1Y1JXW5 A0A084VHY9 A0A182YSJ7 A0A182LH70 A0A1Q3FFB9 B0XD98 R4UMY6 A0A182WXF5 B4M5R0 A0A023EIB7 A0A023EIC3 A0A182IRX3 E2B052 A0A2M3Z2Z7 A0A2M4C0U2 A0A182FBY0 B3P770 A0A2M4A7N4 T1DU70 J3JXQ5 A0A0B4K7R1 Q9VCA9 B3M0J8 B4JH35 A0A1S3DMD4 W5JAG0 B4PKZ4 A0A182MRI0 N6UKB3 A0A0K8TQ55 A0A336KDB6 A0A182PSU3 A0A0K8U9T6 A0A0M4EKV4 A0A182S090 A0A194R9G5 A0A3B0JP43 Q298I2 B4G3Z3 A0A194QC05 A0A182N3A6 A0A034W0F5 A0A0C9QGF2 A0A2J7Q682 A0A1W4V479 A0A182W1P2 B4K7G2 A0A0A1WYM2 D1ZZP0 A0A310SIA3 A0A1J1J6U1 A0A1L8EHJ2 A0A0A9WST4 A0A158N9C9 A0A026WJN9 A0A1W4X1I6 A0A0C9RM97 A0A1I8M4T6 A0A1B0BAA7 D3TNN0 A0A1B0FGJ0 A0A1A9VMX2 A0A1B0A2G5 A0A154P0L1 A0A1W7R9K2 A0A1I8PFR4 A0A2R7WLQ1 A0A1A9W4L2 D3TNM9 A0A087ZPW9 T1JM89 A0A0M8ZRA7 A0A151XFY5
A0A212FP10 S4PYE5 E0VVU3 A0A1B6K719 A0A1B6FD55 U5EZA3 A0A1B6DLS3 T1E333 B4NAG0 A0A1B6KPJ9 A0A1B0EY93 A0A1L8DV91 Q1HRQ4 B4QT28 A0A182Q241 B4HGK6 A0A1B0CYQ9 A0A1S4HE52 A0A2C9GRH5 A0A182USP7 A0A182U3B1 A0A1Y1JXW5 A0A084VHY9 A0A182YSJ7 A0A182LH70 A0A1Q3FFB9 B0XD98 R4UMY6 A0A182WXF5 B4M5R0 A0A023EIB7 A0A023EIC3 A0A182IRX3 E2B052 A0A2M3Z2Z7 A0A2M4C0U2 A0A182FBY0 B3P770 A0A2M4A7N4 T1DU70 J3JXQ5 A0A0B4K7R1 Q9VCA9 B3M0J8 B4JH35 A0A1S3DMD4 W5JAG0 B4PKZ4 A0A182MRI0 N6UKB3 A0A0K8TQ55 A0A336KDB6 A0A182PSU3 A0A0K8U9T6 A0A0M4EKV4 A0A182S090 A0A194R9G5 A0A3B0JP43 Q298I2 B4G3Z3 A0A194QC05 A0A182N3A6 A0A034W0F5 A0A0C9QGF2 A0A2J7Q682 A0A1W4V479 A0A182W1P2 B4K7G2 A0A0A1WYM2 D1ZZP0 A0A310SIA3 A0A1J1J6U1 A0A1L8EHJ2 A0A0A9WST4 A0A158N9C9 A0A026WJN9 A0A1W4X1I6 A0A0C9RM97 A0A1I8M4T6 A0A1B0BAA7 D3TNN0 A0A1B0FGJ0 A0A1A9VMX2 A0A1B0A2G5 A0A154P0L1 A0A1W7R9K2 A0A1I8PFR4 A0A2R7WLQ1 A0A1A9W4L2 D3TNM9 A0A087ZPW9 T1JM89 A0A0M8ZRA7 A0A151XFY5
EC Number
3.4.-.-
Pubmed
22651552
22118469
23622113
20566863
24330624
17994087
+ More
17204158 17510324 12364791 28004739 24438588 25244985 20966253 24945155 26483478 20798317 22516182 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15901661 20920257 23761445 17550304 23537049 26369729 26354079 15632085 25348373 25830018 18362917 19820115 25401762 26823975 21347285 24508170 25315136 20353571
17204158 17510324 12364791 28004739 24438588 25244985 20966253 24945155 26483478 20798317 22516182 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 15901661 20920257 23761445 17550304 23537049 26369729 26354079 15632085 25348373 25830018 18362917 19820115 25401762 26823975 21347285 24508170 25315136 20353571
EMBL
DQ443238
ABF51327.1
ODYU01000577
SOQ35584.1
NWSH01002453
PCG68264.1
+ More
GDQN01001201 JAT89853.1 AK401627 BAM18249.1 RSAL01000074 RVE48961.1 AGBW02004200 OWR55467.1 GAIX01003428 JAA89132.1 DS235814 EEB17499.1 GECU01000459 JAT07248.1 GECZ01021876 JAS47893.1 GANO01001154 JAB58717.1 GEDC01010700 JAS26598.1 GALA01000735 JAA94117.1 CH964232 EDW80774.1 GEBQ01026599 JAT13378.1 AJWK01017654 GFDF01003745 JAV10339.1 DQ440040 CH477204 ABF18073.1 EAT47974.1 CM000364 EDX14192.1 AXCN02001118 CH480815 EDW43459.1 AJVK01009300 AAAB01008816 APCN01004488 GEZM01098148 JAV53973.1 ATLV01013250 KE524847 KFB37583.1 GFDL01008814 JAV26231.1 DS233746 DS232746 EDS31438.1 EDS45352.1 KC571981 AGM32480.1 CH940652 EDW58986.1 GAPW01004928 JAC08670.1 JXUM01145177 GAPW01004930 GAPW01004929 GAPW01004927 KQ569935 JAC08671.1 KXJ68481.1 GL444392 EFN60937.1 GGFM01002146 MBW22897.1 GGFJ01009786 MBW58927.1 CH954182 EDV53890.1 GGFK01003431 MBW36752.1 GAMD01000414 JAB01177.1 BT128026 AEE62987.1 AE014297 AFH06597.1 AY070615 CH902617 EDV44245.1 CH916369 EDV92726.1 ADMH02001689 ETN61422.1 CM000160 EDW98979.1 AXCM01001158 APGK01010787 APGK01029690 KB740739 KB738301 KB632179 ENN79112.1 ENN82676.1 ERL89610.1 GDAI01001332 JAI16271.1 UFQS01000290 UFQT01000290 SSX02489.1 SSX22863.1 GDHF01028993 JAI23321.1 CP012526 ALC47177.1 ALC47908.1 KQ460761 KPJ12511.1 OUUW01000007 SPP82683.1 CM000070 EAL27973.1 CH479179 EDW24404.1 KQ459439 KPJ00961.1 GAKP01011317 JAC47635.1 GBYB01002529 JAG72296.1 NEVH01017535 PNF24088.1 CH933806 EDW14286.1 GBXI01010551 JAD03741.1 KQ971338 EFA02397.1 KQ760832 OAD58889.1 CVRI01000074 CRL08104.1 GFDG01000675 JAV18124.1 GBHO01032092 GDHC01007831 JAG11512.1 JAQ10798.1 ADTU01009386 KK107211 EZA55334.1 GBYB01008101 JAG77868.1 JXJN01010888 EZ423032 ADD19308.1 CCAG010006947 KQ434791 KZC05373.1 GFAH01000567 JAV47822.1 KK855052 PTY20542.1 EZ423031 ADD19307.1 JH432064 KQ435966 KOX67929.1 KQ982174 KYQ59299.1
GDQN01001201 JAT89853.1 AK401627 BAM18249.1 RSAL01000074 RVE48961.1 AGBW02004200 OWR55467.1 GAIX01003428 JAA89132.1 DS235814 EEB17499.1 GECU01000459 JAT07248.1 GECZ01021876 JAS47893.1 GANO01001154 JAB58717.1 GEDC01010700 JAS26598.1 GALA01000735 JAA94117.1 CH964232 EDW80774.1 GEBQ01026599 JAT13378.1 AJWK01017654 GFDF01003745 JAV10339.1 DQ440040 CH477204 ABF18073.1 EAT47974.1 CM000364 EDX14192.1 AXCN02001118 CH480815 EDW43459.1 AJVK01009300 AAAB01008816 APCN01004488 GEZM01098148 JAV53973.1 ATLV01013250 KE524847 KFB37583.1 GFDL01008814 JAV26231.1 DS233746 DS232746 EDS31438.1 EDS45352.1 KC571981 AGM32480.1 CH940652 EDW58986.1 GAPW01004928 JAC08670.1 JXUM01145177 GAPW01004930 GAPW01004929 GAPW01004927 KQ569935 JAC08671.1 KXJ68481.1 GL444392 EFN60937.1 GGFM01002146 MBW22897.1 GGFJ01009786 MBW58927.1 CH954182 EDV53890.1 GGFK01003431 MBW36752.1 GAMD01000414 JAB01177.1 BT128026 AEE62987.1 AE014297 AFH06597.1 AY070615 CH902617 EDV44245.1 CH916369 EDV92726.1 ADMH02001689 ETN61422.1 CM000160 EDW98979.1 AXCM01001158 APGK01010787 APGK01029690 KB740739 KB738301 KB632179 ENN79112.1 ENN82676.1 ERL89610.1 GDAI01001332 JAI16271.1 UFQS01000290 UFQT01000290 SSX02489.1 SSX22863.1 GDHF01028993 JAI23321.1 CP012526 ALC47177.1 ALC47908.1 KQ460761 KPJ12511.1 OUUW01000007 SPP82683.1 CM000070 EAL27973.1 CH479179 EDW24404.1 KQ459439 KPJ00961.1 GAKP01011317 JAC47635.1 GBYB01002529 JAG72296.1 NEVH01017535 PNF24088.1 CH933806 EDW14286.1 GBXI01010551 JAD03741.1 KQ971338 EFA02397.1 KQ760832 OAD58889.1 CVRI01000074 CRL08104.1 GFDG01000675 JAV18124.1 GBHO01032092 GDHC01007831 JAG11512.1 JAQ10798.1 ADTU01009386 KK107211 EZA55334.1 GBYB01008101 JAG77868.1 JXJN01010888 EZ423032 ADD19308.1 CCAG010006947 KQ434791 KZC05373.1 GFAH01000567 JAV47822.1 KK855052 PTY20542.1 EZ423031 ADD19307.1 JH432064 KQ435966 KOX67929.1 KQ982174 KYQ59299.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000009046
UP000007798
UP000092461
+ More
UP000008820 UP000000304 UP000075886 UP000001292 UP000092462 UP000075840 UP000075903 UP000075902 UP000030765 UP000076408 UP000075882 UP000002320 UP000076407 UP000008792 UP000069940 UP000249989 UP000075880 UP000000311 UP000069272 UP000008711 UP000000803 UP000007801 UP000001070 UP000079169 UP000000673 UP000002282 UP000075883 UP000019118 UP000030742 UP000075885 UP000092553 UP000075900 UP000053240 UP000268350 UP000001819 UP000008744 UP000053268 UP000075884 UP000235965 UP000192221 UP000075920 UP000009192 UP000007266 UP000183832 UP000005205 UP000053097 UP000192223 UP000095301 UP000092460 UP000092444 UP000078200 UP000092445 UP000076502 UP000095300 UP000091820 UP000005203 UP000053105 UP000075809
UP000008820 UP000000304 UP000075886 UP000001292 UP000092462 UP000075840 UP000075903 UP000075902 UP000030765 UP000076408 UP000075882 UP000002320 UP000076407 UP000008792 UP000069940 UP000249989 UP000075880 UP000000311 UP000069272 UP000008711 UP000000803 UP000007801 UP000001070 UP000079169 UP000000673 UP000002282 UP000075883 UP000019118 UP000030742 UP000075885 UP000092553 UP000075900 UP000053240 UP000268350 UP000001819 UP000008744 UP000053268 UP000075884 UP000235965 UP000192221 UP000075920 UP000009192 UP000007266 UP000183832 UP000005205 UP000053097 UP000192223 UP000095301 UP000092460 UP000092444 UP000078200 UP000092445 UP000076502 UP000095300 UP000091820 UP000005203 UP000053105 UP000075809
Interpro
Gene 3D
ProteinModelPortal
Q1HQ14
A0A2H1V5P2
A0A2A4J9Q2
A0A1E1WS34
I4DK09
A0A3S2M1P9
+ More
A0A212FP10 S4PYE5 E0VVU3 A0A1B6K719 A0A1B6FD55 U5EZA3 A0A1B6DLS3 T1E333 B4NAG0 A0A1B6KPJ9 A0A1B0EY93 A0A1L8DV91 Q1HRQ4 B4QT28 A0A182Q241 B4HGK6 A0A1B0CYQ9 A0A1S4HE52 A0A2C9GRH5 A0A182USP7 A0A182U3B1 A0A1Y1JXW5 A0A084VHY9 A0A182YSJ7 A0A182LH70 A0A1Q3FFB9 B0XD98 R4UMY6 A0A182WXF5 B4M5R0 A0A023EIB7 A0A023EIC3 A0A182IRX3 E2B052 A0A2M3Z2Z7 A0A2M4C0U2 A0A182FBY0 B3P770 A0A2M4A7N4 T1DU70 J3JXQ5 A0A0B4K7R1 Q9VCA9 B3M0J8 B4JH35 A0A1S3DMD4 W5JAG0 B4PKZ4 A0A182MRI0 N6UKB3 A0A0K8TQ55 A0A336KDB6 A0A182PSU3 A0A0K8U9T6 A0A0M4EKV4 A0A182S090 A0A194R9G5 A0A3B0JP43 Q298I2 B4G3Z3 A0A194QC05 A0A182N3A6 A0A034W0F5 A0A0C9QGF2 A0A2J7Q682 A0A1W4V479 A0A182W1P2 B4K7G2 A0A0A1WYM2 D1ZZP0 A0A310SIA3 A0A1J1J6U1 A0A1L8EHJ2 A0A0A9WST4 A0A158N9C9 A0A026WJN9 A0A1W4X1I6 A0A0C9RM97 A0A1I8M4T6 A0A1B0BAA7 D3TNN0 A0A1B0FGJ0 A0A1A9VMX2 A0A1B0A2G5 A0A154P0L1 A0A1W7R9K2 A0A1I8PFR4 A0A2R7WLQ1 A0A1A9W4L2 D3TNM9 A0A087ZPW9 T1JM89 A0A0M8ZRA7 A0A151XFY5
A0A212FP10 S4PYE5 E0VVU3 A0A1B6K719 A0A1B6FD55 U5EZA3 A0A1B6DLS3 T1E333 B4NAG0 A0A1B6KPJ9 A0A1B0EY93 A0A1L8DV91 Q1HRQ4 B4QT28 A0A182Q241 B4HGK6 A0A1B0CYQ9 A0A1S4HE52 A0A2C9GRH5 A0A182USP7 A0A182U3B1 A0A1Y1JXW5 A0A084VHY9 A0A182YSJ7 A0A182LH70 A0A1Q3FFB9 B0XD98 R4UMY6 A0A182WXF5 B4M5R0 A0A023EIB7 A0A023EIC3 A0A182IRX3 E2B052 A0A2M3Z2Z7 A0A2M4C0U2 A0A182FBY0 B3P770 A0A2M4A7N4 T1DU70 J3JXQ5 A0A0B4K7R1 Q9VCA9 B3M0J8 B4JH35 A0A1S3DMD4 W5JAG0 B4PKZ4 A0A182MRI0 N6UKB3 A0A0K8TQ55 A0A336KDB6 A0A182PSU3 A0A0K8U9T6 A0A0M4EKV4 A0A182S090 A0A194R9G5 A0A3B0JP43 Q298I2 B4G3Z3 A0A194QC05 A0A182N3A6 A0A034W0F5 A0A0C9QGF2 A0A2J7Q682 A0A1W4V479 A0A182W1P2 B4K7G2 A0A0A1WYM2 D1ZZP0 A0A310SIA3 A0A1J1J6U1 A0A1L8EHJ2 A0A0A9WST4 A0A158N9C9 A0A026WJN9 A0A1W4X1I6 A0A0C9RM97 A0A1I8M4T6 A0A1B0BAA7 D3TNN0 A0A1B0FGJ0 A0A1A9VMX2 A0A1B0A2G5 A0A154P0L1 A0A1W7R9K2 A0A1I8PFR4 A0A2R7WLQ1 A0A1A9W4L2 D3TNM9 A0A087ZPW9 T1JM89 A0A0M8ZRA7 A0A151XFY5
Ontologies
GO
PANTHER
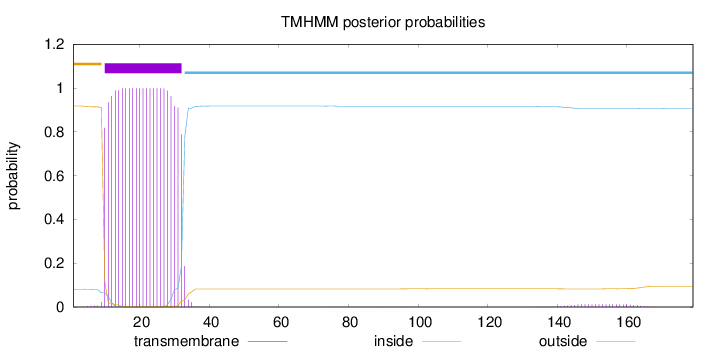
Topology
Subcellular location
Endoplasmic reticulum membrane
Microsome membrane
Microsome membrane
Length:
179
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.82217
Exp number, first 60 AAs:
22.54046
Total prob of N-in:
0.08161
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 179
Population Genetic Test Statistics
Pi
280.950554
Theta
224.452275
Tajima's D
1.01075
CLR
0.192989
CSRT
0.668066596670167
Interpretation
Uncertain
