Gene
KWMTBOMO08010
Pre Gene Modal
BGIBMGA001019
Annotation
PREDICTED:_FGFR1_oncogene_partner_2_homolog_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.498
Sequence
CDS
ATGTCACTGACTATCCAGCAAATAATTTTGGATGCAAAAAGGCTCGCAGGTCGTCTTAAAGAACGCGAAACAGAGGCTGATGCTCTGTTGAGCGAAACCCAAGCAACTTATCGACAAATTCATACAATGAAACAGTATAAAGAAGAAGTTGATACATTAAATGAGGCATCACGAGAGAGACCAAGAGGGGCTCTCATCGCCAGCATAGAGAGAGAGTCTCAGCTCATGAGGGGTGTGCAGAGGGAGAATGGTGAGCTCAGAGCTGCACTAGAAGATCACAGACGAGCCTTGGAATTAATCATGTCAAAATATCGACAGCATACAGAGAAGAAAATATGGGAGTCAAGAATAGATTTCACCTCAGCTATAAATGAGAAACAACAAGAGTTGATACGCCAGCAAGCAGAACGAATCAATGAAATGACAACTGTTATGTACAAAGCAATAAATATGGACGAGAATAGTGAAGCCAGGAAGGATGAAGAGCTTTACCAAAGGCTCATTACTGAAAACAAGGGCCTGAGAGAAATGCTGGATCTATCTCGAAGGTACGGGTCTGATCGTGTGATGGCGCCTCCAATGGAGGACAAGGACGTTCAAACAGACGGCCCGCCGCTGACGGGAGCGTGA
Protein
MSLTIQQIILDAKRLAGRLKERETEADALLSETQATYRQIHTMKQYKEEVDTLNEASRERPRGALIASIERESQLMRGVQRENGELRAALEDHRRALELIMSKYRQHTEKKIWESRIDFTSAINEKQQELIRQQAERINEMTTVMYKAINMDENSEARKDEELYQRLITENKGLREMLDLSRRYGSDRVMAPPMEDKDVQTDGPPLTGA
Summary
Uniprot
H9IUT9
A0A3S2TKY4
A0A2H1V416
I4DP98
A0A194R4A4
A0A194Q7I7
+ More
S4PI27 A0A2A4J923 A0A212FNY7 A0A1W4XAU9 D1ZZP1 A0A1Y1M7X1 A0A1Q3FXT0 A0A1Q3FXQ0 A0A1Q3FXU8 A0A1Q3FXY7 A0A1Q3FXR6 A0A1Q3FY08 A0A336LY14 A0A026WEY1 Q0IG53 A0A1S4F549 A0A023EM66 A0A151IKU2 A0A158N9K1 A0A2R7W708 A0A182SRQ7 A0A151XGG6 F4X7T6 A0A195E1K8 A0A182UQ74 A0A182R9E3 A0A151I4J9 A0A182FBT9 A0A182TNT5 A0A2M4BXR9 A0A182YC79 A0A182MA58 A0A084WM54 E2BNS2 A0A182WD47 A0A0V0GCY7 A0A067RB74 N6UV04 A0A023F995 A0A151JWM8 A0A182KT74 A0A182X051 E9IW69 A0A0J7KVS9 Q7PZW4 A0A182HPM7 A0A182K2X9 A0A2M3ZA42 A0A2M3Z9M0 A0A2M4BTW8 A0A2M4BTX4 N6UES8 A0A182P1G9 A0A182JAM3 J3JYF8 W5JWD7 U5EVB1 A0A182N327 E2AEB3 A0A2M4AS62 A0A2M4AQW1 A0A2M4AR32 A0A182Q2K6 A0A2P8YEY6 A0A2M4ARY4 A0A226E4R0 A0A2A3EJ12 A0A088AQN1 A0A0P4VNI9 T1HJQ3 R4G3Q7 A0A0A9XYV0 A0A0P5RMZ2 A0A162S4D5 A0A0P6II34 A0A310S8X8 A0A310SIJ2 A0A0P6EAS8 T1JNM9 A0A2J7Q674 A0A1S4E7F0 A0A154NZF1 A0A0L7RK66 A0A2M3Z9Q4 A0A2R5L7M6 A0A1D2MX48 A0A293LC43 A0A1Z5L9K9 A0A1Y1M7A0 A0A1I8NGU0 T1PB22 A0A0N7ZDQ3
S4PI27 A0A2A4J923 A0A212FNY7 A0A1W4XAU9 D1ZZP1 A0A1Y1M7X1 A0A1Q3FXT0 A0A1Q3FXQ0 A0A1Q3FXU8 A0A1Q3FXY7 A0A1Q3FXR6 A0A1Q3FY08 A0A336LY14 A0A026WEY1 Q0IG53 A0A1S4F549 A0A023EM66 A0A151IKU2 A0A158N9K1 A0A2R7W708 A0A182SRQ7 A0A151XGG6 F4X7T6 A0A195E1K8 A0A182UQ74 A0A182R9E3 A0A151I4J9 A0A182FBT9 A0A182TNT5 A0A2M4BXR9 A0A182YC79 A0A182MA58 A0A084WM54 E2BNS2 A0A182WD47 A0A0V0GCY7 A0A067RB74 N6UV04 A0A023F995 A0A151JWM8 A0A182KT74 A0A182X051 E9IW69 A0A0J7KVS9 Q7PZW4 A0A182HPM7 A0A182K2X9 A0A2M3ZA42 A0A2M3Z9M0 A0A2M4BTW8 A0A2M4BTX4 N6UES8 A0A182P1G9 A0A182JAM3 J3JYF8 W5JWD7 U5EVB1 A0A182N327 E2AEB3 A0A2M4AS62 A0A2M4AQW1 A0A2M4AR32 A0A182Q2K6 A0A2P8YEY6 A0A2M4ARY4 A0A226E4R0 A0A2A3EJ12 A0A088AQN1 A0A0P4VNI9 T1HJQ3 R4G3Q7 A0A0A9XYV0 A0A0P5RMZ2 A0A162S4D5 A0A0P6II34 A0A310S8X8 A0A310SIJ2 A0A0P6EAS8 T1JNM9 A0A2J7Q674 A0A1S4E7F0 A0A154NZF1 A0A0L7RK66 A0A2M3Z9Q4 A0A2R5L7M6 A0A1D2MX48 A0A293LC43 A0A1Z5L9K9 A0A1Y1M7A0 A0A1I8NGU0 T1PB22 A0A0N7ZDQ3
Pubmed
19121390
22651552
26354079
23622113
22118469
18362917
+ More
19820115 28004739 24508170 30249741 17510324 24945155 21347285 21719571 25244985 24438588 20798317 24845553 23537049 25474469 20966253 21282665 12364791 14747013 17210077 22516182 20920257 23761445 29403074 27129103 25401762 26823975 27289101 28528879 25315136
19820115 28004739 24508170 30249741 17510324 24945155 21347285 21719571 25244985 24438588 20798317 24845553 23537049 25474469 20966253 21282665 12364791 14747013 17210077 22516182 20920257 23761445 29403074 27129103 25401762 26823975 27289101 28528879 25315136
EMBL
BABH01000547
RSAL01000074
RVE48960.1
ODYU01000577
SOQ35585.1
AK403487
+ More
BAM19738.1 KQ460761 KPJ12512.1 KQ459439 KPJ00960.1 GAIX01005570 JAA86990.1 NWSH01002453 PCG68266.1 AGBW02004200 OWR55466.1 KQ971338 EFA01821.1 GEZM01038407 JAV81651.1 GFDL01002675 JAV32370.1 GFDL01002688 JAV32357.1 GFDL01002646 JAV32399.1 GFDL01002672 JAV32373.1 GFDL01002681 JAV32364.1 GFDL01002649 JAV32396.1 UFQT01000290 SSX22864.1 KK107260 QOIP01000001 EZA54231.1 RLU26785.1 CH477274 EAT45158.1 GAPW01003212 JAC10386.1 KQ977203 KYN04967.1 ADTU01009678 KK854269 PTY13995.1 KQ982169 KYQ59486.1 GL888891 EGI57492.1 KQ979814 KYN19035.1 KQ976461 KYM84686.1 GGFJ01008708 MBW57849.1 AXCM01004725 ATLV01024388 KE525351 KFB51298.1 GL449462 EFN82655.1 GECL01000145 JAP05979.1 KK852761 KDR17017.1 APGK01010788 KB738301 KB632179 ENN82677.1 ERL89611.1 GBBI01001143 JAC17569.1 KQ981629 KYN38966.1 GL766449 EFZ15203.1 LBMM01002628 KMQ94572.1 AAAB01008986 EAA00216.3 APCN01003265 GGFM01004594 MBW25345.1 GGFM01004486 MBW25237.1 GGFJ01007313 MBW56454.1 GGFJ01007311 MBW56452.1 APGK01029689 KB740739 ENN79111.1 BT128284 AEE63244.1 ADMH02000225 ETN67304.1 GANO01003485 JAB56386.1 GL438827 EFN68346.1 GGFK01010107 MBW43428.1 GGFK01009855 MBW43176.1 GGFK01009909 MBW43230.1 AXCN02001131 PYGN01000650 PSN42827.1 GGFK01010208 MBW43529.1 LNIX01000006 OXA52725.1 KZ288253 PBC31011.1 GDKW01002534 JAI54061.1 ACPB03021765 ACPB03021766 GAHY01002010 JAA75500.1 GBHO01019572 GBHO01019571 GBRD01010098 GBRD01010096 GBRD01010095 GDHC01020655 GDHC01003775 JAG24032.1 JAG24033.1 JAG55726.1 JAP97973.1 JAQ14854.1 GDIQ01098463 JAL53263.1 LRGB01000084 KZS21004.1 GDIQ01010682 JAN84055.1 KQ769580 OAD52851.1 KQ762400 OAD55890.1 GDIQ01071075 JAN23662.1 JH432114 NEVH01017535 PNF24090.1 KQ434777 KZC04250.1 KQ414578 KOC71203.1 GGFM01004503 MBW25254.1 GGLE01001356 MBY05482.1 LJIJ01000459 ODM97255.1 GFWV01000836 MAA25566.1 GFJQ02002857 JAW04113.1 GEZM01038406 JAV81652.1 KA645153 AFP59782.1 GDRN01011171 JAI67888.1
BAM19738.1 KQ460761 KPJ12512.1 KQ459439 KPJ00960.1 GAIX01005570 JAA86990.1 NWSH01002453 PCG68266.1 AGBW02004200 OWR55466.1 KQ971338 EFA01821.1 GEZM01038407 JAV81651.1 GFDL01002675 JAV32370.1 GFDL01002688 JAV32357.1 GFDL01002646 JAV32399.1 GFDL01002672 JAV32373.1 GFDL01002681 JAV32364.1 GFDL01002649 JAV32396.1 UFQT01000290 SSX22864.1 KK107260 QOIP01000001 EZA54231.1 RLU26785.1 CH477274 EAT45158.1 GAPW01003212 JAC10386.1 KQ977203 KYN04967.1 ADTU01009678 KK854269 PTY13995.1 KQ982169 KYQ59486.1 GL888891 EGI57492.1 KQ979814 KYN19035.1 KQ976461 KYM84686.1 GGFJ01008708 MBW57849.1 AXCM01004725 ATLV01024388 KE525351 KFB51298.1 GL449462 EFN82655.1 GECL01000145 JAP05979.1 KK852761 KDR17017.1 APGK01010788 KB738301 KB632179 ENN82677.1 ERL89611.1 GBBI01001143 JAC17569.1 KQ981629 KYN38966.1 GL766449 EFZ15203.1 LBMM01002628 KMQ94572.1 AAAB01008986 EAA00216.3 APCN01003265 GGFM01004594 MBW25345.1 GGFM01004486 MBW25237.1 GGFJ01007313 MBW56454.1 GGFJ01007311 MBW56452.1 APGK01029689 KB740739 ENN79111.1 BT128284 AEE63244.1 ADMH02000225 ETN67304.1 GANO01003485 JAB56386.1 GL438827 EFN68346.1 GGFK01010107 MBW43428.1 GGFK01009855 MBW43176.1 GGFK01009909 MBW43230.1 AXCN02001131 PYGN01000650 PSN42827.1 GGFK01010208 MBW43529.1 LNIX01000006 OXA52725.1 KZ288253 PBC31011.1 GDKW01002534 JAI54061.1 ACPB03021765 ACPB03021766 GAHY01002010 JAA75500.1 GBHO01019572 GBHO01019571 GBRD01010098 GBRD01010096 GBRD01010095 GDHC01020655 GDHC01003775 JAG24032.1 JAG24033.1 JAG55726.1 JAP97973.1 JAQ14854.1 GDIQ01098463 JAL53263.1 LRGB01000084 KZS21004.1 GDIQ01010682 JAN84055.1 KQ769580 OAD52851.1 KQ762400 OAD55890.1 GDIQ01071075 JAN23662.1 JH432114 NEVH01017535 PNF24090.1 KQ434777 KZC04250.1 KQ414578 KOC71203.1 GGFM01004503 MBW25254.1 GGLE01001356 MBY05482.1 LJIJ01000459 ODM97255.1 GFWV01000836 MAA25566.1 GFJQ02002857 JAW04113.1 GEZM01038406 JAV81652.1 KA645153 AFP59782.1 GDRN01011171 JAI67888.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000218220
UP000007151
+ More
UP000192223 UP000007266 UP000053097 UP000279307 UP000008820 UP000078542 UP000005205 UP000075901 UP000075809 UP000007755 UP000078492 UP000075903 UP000075900 UP000078540 UP000069272 UP000075902 UP000076408 UP000075883 UP000030765 UP000008237 UP000075920 UP000027135 UP000019118 UP000030742 UP000078541 UP000075882 UP000076407 UP000036403 UP000007062 UP000075840 UP000075881 UP000075885 UP000075880 UP000000673 UP000075884 UP000000311 UP000075886 UP000245037 UP000198287 UP000242457 UP000005203 UP000015103 UP000076858 UP000235965 UP000079169 UP000076502 UP000053825 UP000094527 UP000095301
UP000192223 UP000007266 UP000053097 UP000279307 UP000008820 UP000078542 UP000005205 UP000075901 UP000075809 UP000007755 UP000078492 UP000075903 UP000075900 UP000078540 UP000069272 UP000075902 UP000076408 UP000075883 UP000030765 UP000008237 UP000075920 UP000027135 UP000019118 UP000030742 UP000078541 UP000075882 UP000076407 UP000036403 UP000007062 UP000075840 UP000075881 UP000075885 UP000075880 UP000000673 UP000075884 UP000000311 UP000075886 UP000245037 UP000198287 UP000242457 UP000005203 UP000015103 UP000076858 UP000235965 UP000079169 UP000076502 UP000053825 UP000094527 UP000095301
PRIDE
Pfam
PF05769 SIKE
Interpro
IPR008555
SIKE
ProteinModelPortal
H9IUT9
A0A3S2TKY4
A0A2H1V416
I4DP98
A0A194R4A4
A0A194Q7I7
+ More
S4PI27 A0A2A4J923 A0A212FNY7 A0A1W4XAU9 D1ZZP1 A0A1Y1M7X1 A0A1Q3FXT0 A0A1Q3FXQ0 A0A1Q3FXU8 A0A1Q3FXY7 A0A1Q3FXR6 A0A1Q3FY08 A0A336LY14 A0A026WEY1 Q0IG53 A0A1S4F549 A0A023EM66 A0A151IKU2 A0A158N9K1 A0A2R7W708 A0A182SRQ7 A0A151XGG6 F4X7T6 A0A195E1K8 A0A182UQ74 A0A182R9E3 A0A151I4J9 A0A182FBT9 A0A182TNT5 A0A2M4BXR9 A0A182YC79 A0A182MA58 A0A084WM54 E2BNS2 A0A182WD47 A0A0V0GCY7 A0A067RB74 N6UV04 A0A023F995 A0A151JWM8 A0A182KT74 A0A182X051 E9IW69 A0A0J7KVS9 Q7PZW4 A0A182HPM7 A0A182K2X9 A0A2M3ZA42 A0A2M3Z9M0 A0A2M4BTW8 A0A2M4BTX4 N6UES8 A0A182P1G9 A0A182JAM3 J3JYF8 W5JWD7 U5EVB1 A0A182N327 E2AEB3 A0A2M4AS62 A0A2M4AQW1 A0A2M4AR32 A0A182Q2K6 A0A2P8YEY6 A0A2M4ARY4 A0A226E4R0 A0A2A3EJ12 A0A088AQN1 A0A0P4VNI9 T1HJQ3 R4G3Q7 A0A0A9XYV0 A0A0P5RMZ2 A0A162S4D5 A0A0P6II34 A0A310S8X8 A0A310SIJ2 A0A0P6EAS8 T1JNM9 A0A2J7Q674 A0A1S4E7F0 A0A154NZF1 A0A0L7RK66 A0A2M3Z9Q4 A0A2R5L7M6 A0A1D2MX48 A0A293LC43 A0A1Z5L9K9 A0A1Y1M7A0 A0A1I8NGU0 T1PB22 A0A0N7ZDQ3
S4PI27 A0A2A4J923 A0A212FNY7 A0A1W4XAU9 D1ZZP1 A0A1Y1M7X1 A0A1Q3FXT0 A0A1Q3FXQ0 A0A1Q3FXU8 A0A1Q3FXY7 A0A1Q3FXR6 A0A1Q3FY08 A0A336LY14 A0A026WEY1 Q0IG53 A0A1S4F549 A0A023EM66 A0A151IKU2 A0A158N9K1 A0A2R7W708 A0A182SRQ7 A0A151XGG6 F4X7T6 A0A195E1K8 A0A182UQ74 A0A182R9E3 A0A151I4J9 A0A182FBT9 A0A182TNT5 A0A2M4BXR9 A0A182YC79 A0A182MA58 A0A084WM54 E2BNS2 A0A182WD47 A0A0V0GCY7 A0A067RB74 N6UV04 A0A023F995 A0A151JWM8 A0A182KT74 A0A182X051 E9IW69 A0A0J7KVS9 Q7PZW4 A0A182HPM7 A0A182K2X9 A0A2M3ZA42 A0A2M3Z9M0 A0A2M4BTW8 A0A2M4BTX4 N6UES8 A0A182P1G9 A0A182JAM3 J3JYF8 W5JWD7 U5EVB1 A0A182N327 E2AEB3 A0A2M4AS62 A0A2M4AQW1 A0A2M4AR32 A0A182Q2K6 A0A2P8YEY6 A0A2M4ARY4 A0A226E4R0 A0A2A3EJ12 A0A088AQN1 A0A0P4VNI9 T1HJQ3 R4G3Q7 A0A0A9XYV0 A0A0P5RMZ2 A0A162S4D5 A0A0P6II34 A0A310S8X8 A0A310SIJ2 A0A0P6EAS8 T1JNM9 A0A2J7Q674 A0A1S4E7F0 A0A154NZF1 A0A0L7RK66 A0A2M3Z9Q4 A0A2R5L7M6 A0A1D2MX48 A0A293LC43 A0A1Z5L9K9 A0A1Y1M7A0 A0A1I8NGU0 T1PB22 A0A0N7ZDQ3
Ontologies
GO
PANTHER
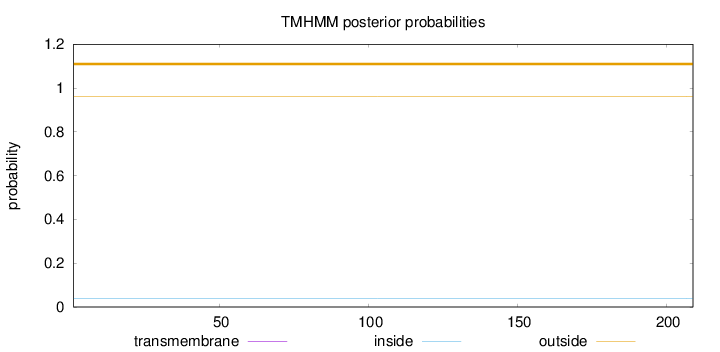
Topology
Length:
209
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03990
outside
1 - 209
Population Genetic Test Statistics
Pi
161.118587
Theta
144.396803
Tajima's D
0.701003
CLR
0
CSRT
0.573971301434928
Interpretation
Uncertain
