Gene
KWMTBOMO08009
Pre Gene Modal
BGIBMGA001018
Annotation
PREDICTED:_epoxide_hydrolase_3-like_[Papilio_xuthus]
Full name
Epoxide hydrolase 4
Alternative Name
Abhydrolase domain-containing protein 7
Epoxide hydrolase-related protein
Epoxide hydrolase-related protein
Location in the cell
PlasmaMembrane Reliability : 2.235
Sequence
CDS
ATGAAGCAATATATTTATCGGTTTGTCGTAACGAACGCGTTAACGCTGTTTTACGGCTTTCTAACTTTAGTGATCTTTCTTCGGAATTATGTAAGGAACCCGTTCCGAAATCCATGGGAGCAGAAGTTGAGGCTTGTCCCGCCCGCGCCTCTATCCGACCCCAAGTATGGAATTCACAAGAATATCAAAGTTAATGGCATAAAACTACATTACGTTGAAAGCGGCGACCCTTCGAAGCCTTTAATGATTTTTCTACATGGTTTTCCTGAATTCTGGTATTCATGGAGATACCAAATTTTGGAATTTAAGAAAGATTATTGGTGCATAGCCGTTGACATGCGAGGATACGGAGATTCCGAAAGAGTTGAAGATGTCTCCGCCTACAAGTTAAATCTGCTCATTGACGACATCAAATATTTAGTTAAGGAGCTGGGTCGTGAGAAGTGCATTTTAGTCTCGCATGACTGGGGAGGTGCGATAGCGAATGCTATACGAAATTCGTACCCCGAAATAGTCAGCGCATTGATTATGTTAGCCAGCATGTCAACCGCAGCTTCTATTCACGAAATATGGAACAATTCAAAGCAGTTCTTAATGTCTTGGTACTTCTTCCTGTTTCGTCTACCTTGGCTGCCAGAGCAGTTTGTCTCAATGAACGACCTTGAGATACATGATAAGATATTACTTGTTCCTGGCAACACGTATGTCGACAAGCAAGATGTCGAATGTTACAAGTACTGGTTTTGTAAGCCTTATGCTCTGACTCCACCCATTAATTACTACCGTGCCAACTTCGATTATATCGCAAATGATAAATATTACAAAGATAATGTTCCAATGCTAGTCGTACACGCTGCCAAAGATTTGTACATCAACAAAACAGTTCTGGAAGCTATGAAGAAACAATACGAATACATCGAAACTGTAATAGTCGACGGTGTCGGACATGCACTACAGCAACACGATCCAGACAGAATCAATAAAATAATTCGAGATTTTCTAGCCAAGAACAATCTATAA
Protein
MKQYIYRFVVTNALTLFYGFLTLVIFLRNYVRNPFRNPWEQKLRLVPPAPLSDPKYGIHKNIKVNGIKLHYVESGDPSKPLMIFLHGFPEFWYSWRYQILEFKKDYWCIAVDMRGYGDSERVEDVSAYKLNLLIDDIKYLVKELGREKCILVSHDWGGAIANAIRNSYPEIVSALIMLASMSTAASIHEIWNNSKQFLMSWYFFLFRLPWLPEQFVSMNDLEIHDKILLVPGNTYVDKQDVECYKYWFCKPYALTPPINYYRANFDYIANDKYYKDNVPMLVVHAAKDLYINKTVLEAMKKQYEYIETVIVDGVGHALQQHDPDRINKIIRDFLAKNNL
Summary
Similarity
Belongs to the AB hydrolase superfamily. Epoxide hydrolase family.
Keywords
Complete proteome
Hydrolase
Membrane
Polymorphism
Reference proteome
Signal-anchor
Transmembrane
Transmembrane helix
Feature
chain Epoxide hydrolase 4
sequence variant In dbSNP:rs17854127.
sequence variant In dbSNP:rs17854127.
Uniprot
H9IUT8
A0A2H1V482
A0A2A4J813
A0A212FP09
A0A3S2P035
A0A194Q6F7
+ More
A0A194R4S5 A0A182JK48 A0A182GRL0 Q176I8 A0A182G6C7 A0A1B6FEJ8 A0A194R3X9 A0A182SM00 A0A182QH98 U5EGL8 Q7PZL6 A0A182MDM7 A0A182U932 A0A182USP8 A0A2M3Z6C4 A0A2M4BT77 A0A2M3Z5K0 A0A2M3Z5J1 A0A2M4BT67 A0A023EQ13 W5JL78 A0A2M4A5K2 B0XJ28 A0A182W230 A0A182YBN1 A0A1Q3FBJ6 A0A182RKG1 A0A2J7R397 A0A182XJM6 A0A067RU58 A0A182FTQ6 A0A182NZY4 A0A182GSE2 Q176I7 A0A067RWQ5 A0A2J7R3C2 A0A336LSA0 B0XJ30 A0A182GRK8 B0WFT3 A0A146KN80 V4BCL3 A0A2J7R388 Q176J0 A0A1B0GL84 A0A067RMS1 A0A1S3HRG0 A0A226ELK1 A0A1B6L515 B0WFT1 A0A1B6JJR5 A0A1L8DYW4 A0A1L8DZB6 A0A0B6YQU2 A0A1B6E5K7 V4BHS9 A0A1J1IC92 A0A1X7U8B3 A0A2C9K8K1 A0A2P8ZI48 A0A0B7B8T7 A0A2M4A7L1 A0A3R7PHY9 A0A0K8SZ58 A0A0A9YNM3 R4G3W1 G3WEW4 T1IWN7 A0A1E1X894 A0A1W0W9R3 A0A0B7BIC4 A0A131Y1H6 A0A099ZQ59 A0A0B6ZKI8 A0A1S3J531 W4Z450 F7G315 A0A0B7B4K1 A0A1V4JYW0 K7F3S7 H2N6S6 A0A0K8RRD7 H2ZVN0 A0A2K5R6M7 F7D2S6 A0A2K6U1F0 G3QD12 G1RHG6 Q8IUS5 A0A0B7B2E3 A0A0Q3MI06 R7TRZ4 A0A2J8T701
A0A194R4S5 A0A182JK48 A0A182GRL0 Q176I8 A0A182G6C7 A0A1B6FEJ8 A0A194R3X9 A0A182SM00 A0A182QH98 U5EGL8 Q7PZL6 A0A182MDM7 A0A182U932 A0A182USP8 A0A2M3Z6C4 A0A2M4BT77 A0A2M3Z5K0 A0A2M3Z5J1 A0A2M4BT67 A0A023EQ13 W5JL78 A0A2M4A5K2 B0XJ28 A0A182W230 A0A182YBN1 A0A1Q3FBJ6 A0A182RKG1 A0A2J7R397 A0A182XJM6 A0A067RU58 A0A182FTQ6 A0A182NZY4 A0A182GSE2 Q176I7 A0A067RWQ5 A0A2J7R3C2 A0A336LSA0 B0XJ30 A0A182GRK8 B0WFT3 A0A146KN80 V4BCL3 A0A2J7R388 Q176J0 A0A1B0GL84 A0A067RMS1 A0A1S3HRG0 A0A226ELK1 A0A1B6L515 B0WFT1 A0A1B6JJR5 A0A1L8DYW4 A0A1L8DZB6 A0A0B6YQU2 A0A1B6E5K7 V4BHS9 A0A1J1IC92 A0A1X7U8B3 A0A2C9K8K1 A0A2P8ZI48 A0A0B7B8T7 A0A2M4A7L1 A0A3R7PHY9 A0A0K8SZ58 A0A0A9YNM3 R4G3W1 G3WEW4 T1IWN7 A0A1E1X894 A0A1W0W9R3 A0A0B7BIC4 A0A131Y1H6 A0A099ZQ59 A0A0B6ZKI8 A0A1S3J531 W4Z450 F7G315 A0A0B7B4K1 A0A1V4JYW0 K7F3S7 H2N6S6 A0A0K8RRD7 H2ZVN0 A0A2K5R6M7 F7D2S6 A0A2K6U1F0 G3QD12 G1RHG6 Q8IUS5 A0A0B7B2E3 A0A0Q3MI06 R7TRZ4 A0A2J8T701
EC Number
3.3.-.-
Pubmed
EMBL
BABH01000547
BABH01000548
ODYU01000577
SOQ35586.1
NWSH01002453
PCG68267.1
+ More
AGBW02004200 OWR55465.1 RSAL01000074 RVE48959.1 KQ459439 KPJ00959.1 KQ460761 KPJ12514.1 JXUM01016362 KQ560456 KXJ82329.1 CH477386 EAT42095.1 JXUM01147659 JXUM01147660 JXUM01147661 KQ570412 KXJ68369.1 GECZ01021152 JAS48617.1 KPJ12513.1 AXCN02001144 GANO01003390 JAB56481.1 AAAB01008986 EAA00354.3 AXCM01000173 GGFM01003325 MBW24076.1 GGFJ01007052 MBW56193.1 GGFM01003041 MBW23792.1 GGFM01003038 MBW23789.1 GGFJ01007053 MBW56194.1 GAPW01002468 JAC11130.1 ADMH02001207 ETN63670.1 GGFK01002773 MBW36094.1 DS233427 EDS29923.1 GFDL01010099 JAV24946.1 NEVH01007822 PNF35321.1 KK852417 KDR24345.1 JXUM01084364 KQ563433 KXJ73838.1 EAT42096.1 KDR24344.1 PNF35320.1 UFQT01000147 SSX20892.1 EDS29925.1 JXUM01016357 KXJ82327.1 DS231920 EDS26427.1 GDHC01021090 JAP97538.1 KB199650 ESP05421.1 PNF35311.1 EAT42093.1 AJWK01034520 KDR24343.1 LNIX01000003 OXA58563.1 GEBQ01021169 GEBQ01006444 JAT18808.1 JAT33533.1 EDS26425.1 GECU01008354 JAS99352.1 GFDF01002458 JAV11626.1 GFDF01002459 JAV11625.1 HACG01011620 HACG01011621 HACG01011622 CEK58485.1 CEK58486.1 CEK58487.1 GEDC01004087 JAS33211.1 ESP05422.1 CVRI01000044 CRK96598.1 PYGN01000050 PSN56166.1 HACG01042462 CEK89327.1 GGFK01003411 MBW36732.1 QCYY01002845 ROT67142.1 GBRD01007312 JAG58509.1 GBHO01010363 JAG33241.1 ACPB03009260 GAHY01001321 JAA76189.1 AEFK01152852 AEFK01152853 JH431625 GFAC01003744 JAT95444.1 MTYJ01000159 OQV11892.1 HACG01045196 CEK92061.1 GEFM01003448 JAP72348.1 KL895887 KGL82920.1 HACG01022279 HACG01022280 CEK69144.1 CEK69145.1 AAGJ04074142 HACG01040165 CEK87030.1 LSYS01005497 OPJ77331.1 AGCU01191113 AGCU01191114 AGCU01191115 AGCU01191116 AGCU01191117 AGCU01191118 ABGA01060702 ABGA01060703 NDHI03003423 PNJ55387.1 GADI01000161 JAA73647.1 AFYH01117506 AFYH01117507 AFYH01117508 AFYH01117509 AFYH01117510 AFYH01117511 AFYH01117512 GAMT01006025 GAMR01010037 GAMP01003144 JAB05836.1 JAB23895.1 JAB49611.1 CABD030004544 ADFV01189711 ADFV01189712 AK074822 BC041475 HACG01040167 CEK87032.1 LMAW01002002 KQK82111.1 AMQN01011323 KB308827 ELT96409.1 NDHI03003519 PNJ28817.1 PNJ28819.1 PNJ28820.1
AGBW02004200 OWR55465.1 RSAL01000074 RVE48959.1 KQ459439 KPJ00959.1 KQ460761 KPJ12514.1 JXUM01016362 KQ560456 KXJ82329.1 CH477386 EAT42095.1 JXUM01147659 JXUM01147660 JXUM01147661 KQ570412 KXJ68369.1 GECZ01021152 JAS48617.1 KPJ12513.1 AXCN02001144 GANO01003390 JAB56481.1 AAAB01008986 EAA00354.3 AXCM01000173 GGFM01003325 MBW24076.1 GGFJ01007052 MBW56193.1 GGFM01003041 MBW23792.1 GGFM01003038 MBW23789.1 GGFJ01007053 MBW56194.1 GAPW01002468 JAC11130.1 ADMH02001207 ETN63670.1 GGFK01002773 MBW36094.1 DS233427 EDS29923.1 GFDL01010099 JAV24946.1 NEVH01007822 PNF35321.1 KK852417 KDR24345.1 JXUM01084364 KQ563433 KXJ73838.1 EAT42096.1 KDR24344.1 PNF35320.1 UFQT01000147 SSX20892.1 EDS29925.1 JXUM01016357 KXJ82327.1 DS231920 EDS26427.1 GDHC01021090 JAP97538.1 KB199650 ESP05421.1 PNF35311.1 EAT42093.1 AJWK01034520 KDR24343.1 LNIX01000003 OXA58563.1 GEBQ01021169 GEBQ01006444 JAT18808.1 JAT33533.1 EDS26425.1 GECU01008354 JAS99352.1 GFDF01002458 JAV11626.1 GFDF01002459 JAV11625.1 HACG01011620 HACG01011621 HACG01011622 CEK58485.1 CEK58486.1 CEK58487.1 GEDC01004087 JAS33211.1 ESP05422.1 CVRI01000044 CRK96598.1 PYGN01000050 PSN56166.1 HACG01042462 CEK89327.1 GGFK01003411 MBW36732.1 QCYY01002845 ROT67142.1 GBRD01007312 JAG58509.1 GBHO01010363 JAG33241.1 ACPB03009260 GAHY01001321 JAA76189.1 AEFK01152852 AEFK01152853 JH431625 GFAC01003744 JAT95444.1 MTYJ01000159 OQV11892.1 HACG01045196 CEK92061.1 GEFM01003448 JAP72348.1 KL895887 KGL82920.1 HACG01022279 HACG01022280 CEK69144.1 CEK69145.1 AAGJ04074142 HACG01040165 CEK87030.1 LSYS01005497 OPJ77331.1 AGCU01191113 AGCU01191114 AGCU01191115 AGCU01191116 AGCU01191117 AGCU01191118 ABGA01060702 ABGA01060703 NDHI03003423 PNJ55387.1 GADI01000161 JAA73647.1 AFYH01117506 AFYH01117507 AFYH01117508 AFYH01117509 AFYH01117510 AFYH01117511 AFYH01117512 GAMT01006025 GAMR01010037 GAMP01003144 JAB05836.1 JAB23895.1 JAB49611.1 CABD030004544 ADFV01189711 ADFV01189712 AK074822 BC041475 HACG01040167 CEK87032.1 LMAW01002002 KQK82111.1 AMQN01011323 KB308827 ELT96409.1 NDHI03003519 PNJ28817.1 PNJ28819.1 PNJ28820.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000075880 UP000069940 UP000249989 UP000008820 UP000075901 UP000075886 UP000007062 UP000075883 UP000075902 UP000075903 UP000000673 UP000002320 UP000075920 UP000076408 UP000075900 UP000235965 UP000076407 UP000027135 UP000069272 UP000075885 UP000030746 UP000092461 UP000085678 UP000198287 UP000183832 UP000007879 UP000076420 UP000245037 UP000283509 UP000015103 UP000007648 UP000053641 UP000007110 UP000002280 UP000190648 UP000007267 UP000001595 UP000008672 UP000233040 UP000008225 UP000233220 UP000001519 UP000001073 UP000005640 UP000051836 UP000014760
UP000075880 UP000069940 UP000249989 UP000008820 UP000075901 UP000075886 UP000007062 UP000075883 UP000075902 UP000075903 UP000000673 UP000002320 UP000075920 UP000076408 UP000075900 UP000235965 UP000076407 UP000027135 UP000069272 UP000075885 UP000030746 UP000092461 UP000085678 UP000198287 UP000183832 UP000007879 UP000076420 UP000245037 UP000283509 UP000015103 UP000007648 UP000053641 UP000007110 UP000002280 UP000190648 UP000007267 UP000001595 UP000008672 UP000233040 UP000008225 UP000233220 UP000001519 UP000001073 UP000005640 UP000051836 UP000014760
Interpro
IPR000639
Epox_hydrolase-like
+ More
IPR000073 AB_hydrolase_1
IPR029058 AB_hydrolase
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR006179 5_nucleotidase/apyrase
IPR029052 Metallo-depent_PP-like
IPR008334 5'-Nucleotdase_C
IPR004843 Calcineurin-like_PHP_ApaH
IPR036907 5'-Nucleotdase_C_sf
IPR006146 5'-Nucleotdase_CS
IPR000073 AB_hydrolase_1
IPR029058 AB_hydrolase
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR006179 5_nucleotidase/apyrase
IPR029052 Metallo-depent_PP-like
IPR008334 5'-Nucleotdase_C
IPR004843 Calcineurin-like_PHP_ApaH
IPR036907 5'-Nucleotdase_C_sf
IPR006146 5'-Nucleotdase_CS
Gene 3D
ProteinModelPortal
H9IUT8
A0A2H1V482
A0A2A4J813
A0A212FP09
A0A3S2P035
A0A194Q6F7
+ More
A0A194R4S5 A0A182JK48 A0A182GRL0 Q176I8 A0A182G6C7 A0A1B6FEJ8 A0A194R3X9 A0A182SM00 A0A182QH98 U5EGL8 Q7PZL6 A0A182MDM7 A0A182U932 A0A182USP8 A0A2M3Z6C4 A0A2M4BT77 A0A2M3Z5K0 A0A2M3Z5J1 A0A2M4BT67 A0A023EQ13 W5JL78 A0A2M4A5K2 B0XJ28 A0A182W230 A0A182YBN1 A0A1Q3FBJ6 A0A182RKG1 A0A2J7R397 A0A182XJM6 A0A067RU58 A0A182FTQ6 A0A182NZY4 A0A182GSE2 Q176I7 A0A067RWQ5 A0A2J7R3C2 A0A336LSA0 B0XJ30 A0A182GRK8 B0WFT3 A0A146KN80 V4BCL3 A0A2J7R388 Q176J0 A0A1B0GL84 A0A067RMS1 A0A1S3HRG0 A0A226ELK1 A0A1B6L515 B0WFT1 A0A1B6JJR5 A0A1L8DYW4 A0A1L8DZB6 A0A0B6YQU2 A0A1B6E5K7 V4BHS9 A0A1J1IC92 A0A1X7U8B3 A0A2C9K8K1 A0A2P8ZI48 A0A0B7B8T7 A0A2M4A7L1 A0A3R7PHY9 A0A0K8SZ58 A0A0A9YNM3 R4G3W1 G3WEW4 T1IWN7 A0A1E1X894 A0A1W0W9R3 A0A0B7BIC4 A0A131Y1H6 A0A099ZQ59 A0A0B6ZKI8 A0A1S3J531 W4Z450 F7G315 A0A0B7B4K1 A0A1V4JYW0 K7F3S7 H2N6S6 A0A0K8RRD7 H2ZVN0 A0A2K5R6M7 F7D2S6 A0A2K6U1F0 G3QD12 G1RHG6 Q8IUS5 A0A0B7B2E3 A0A0Q3MI06 R7TRZ4 A0A2J8T701
A0A194R4S5 A0A182JK48 A0A182GRL0 Q176I8 A0A182G6C7 A0A1B6FEJ8 A0A194R3X9 A0A182SM00 A0A182QH98 U5EGL8 Q7PZL6 A0A182MDM7 A0A182U932 A0A182USP8 A0A2M3Z6C4 A0A2M4BT77 A0A2M3Z5K0 A0A2M3Z5J1 A0A2M4BT67 A0A023EQ13 W5JL78 A0A2M4A5K2 B0XJ28 A0A182W230 A0A182YBN1 A0A1Q3FBJ6 A0A182RKG1 A0A2J7R397 A0A182XJM6 A0A067RU58 A0A182FTQ6 A0A182NZY4 A0A182GSE2 Q176I7 A0A067RWQ5 A0A2J7R3C2 A0A336LSA0 B0XJ30 A0A182GRK8 B0WFT3 A0A146KN80 V4BCL3 A0A2J7R388 Q176J0 A0A1B0GL84 A0A067RMS1 A0A1S3HRG0 A0A226ELK1 A0A1B6L515 B0WFT1 A0A1B6JJR5 A0A1L8DYW4 A0A1L8DZB6 A0A0B6YQU2 A0A1B6E5K7 V4BHS9 A0A1J1IC92 A0A1X7U8B3 A0A2C9K8K1 A0A2P8ZI48 A0A0B7B8T7 A0A2M4A7L1 A0A3R7PHY9 A0A0K8SZ58 A0A0A9YNM3 R4G3W1 G3WEW4 T1IWN7 A0A1E1X894 A0A1W0W9R3 A0A0B7BIC4 A0A131Y1H6 A0A099ZQ59 A0A0B6ZKI8 A0A1S3J531 W4Z450 F7G315 A0A0B7B4K1 A0A1V4JYW0 K7F3S7 H2N6S6 A0A0K8RRD7 H2ZVN0 A0A2K5R6M7 F7D2S6 A0A2K6U1F0 G3QD12 G1RHG6 Q8IUS5 A0A0B7B2E3 A0A0Q3MI06 R7TRZ4 A0A2J8T701
PDB
5NG7
E-value=3.80022e-39,
Score=405
Ontologies
GO
PANTHER
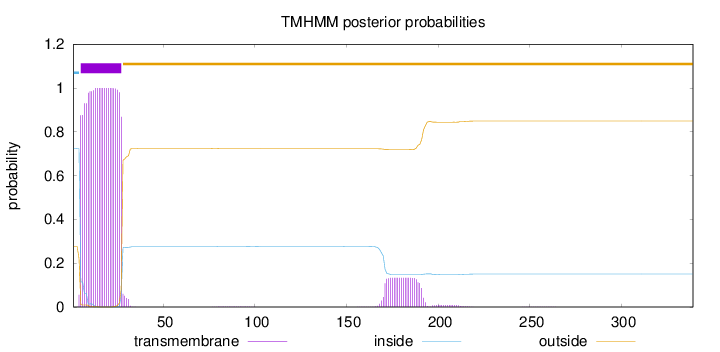
Topology
Subcellular location
Membrane
Length:
339
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.66235
Exp number, first 60 AAs:
22.63307
Total prob of N-in:
0.72398
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 339
Population Genetic Test Statistics
Pi
221.074281
Theta
168.449168
Tajima's D
1.216467
CLR
0.037554
CSRT
0.71746412679366
Interpretation
Uncertain
