Pre Gene Modal
BGIBMGA001137
Annotation
thymidylate_synthase_isoform_1_[Bombyx_mori]
Full name
Thymidylate synthase
Location in the cell
Cytoplasmic Reliability : 2.19
Sequence
CDS
ATGACACACAATAACGATGTCGAAATAAAGTCTCATGATGAATATCAATATTTAAAACTTATCGACCAAATCATAAAATCTGGCGATAAAAGAGTTGATAGAACGGGGGTAGGTACCCTTTCGATATTTGGAGCGATGCAGAGGTACTCTCTTCACAATAACACGCTTCCACTTCTCACAACTAAACGAGTATTTACAAGAGGTGTTATAGAAGAACTACTGTGGATGATCTCCGGGTCTACGGATTCAAAAGCATTAGCTGCCAAAGGTGTACACATTTGGGATGCAAATGGATCAAGAAATTTCCTTGACAATTTAGGATTTACTGACAGAGAAGAAGGAGATCTAGGCCCTGTATATGGATTTCAATGGAGACATAGTGGTGCAAAATATATTGATTGTCAGACTGATTATACTGGACAGGGTATAGACCAACTGCAAAGTGTAATAGATTCTATTAAGAACAATCCAGCAGACAGAAGAATGTTGATATGTGCATGGAATTCTTCAGACCTTAAGAAGATGGCACTACCACCATGTCACTGTTTAGCACAATTCTATGTGGCTGATGGGAAATTATCTTGCTTGTTGTACCAAAGAAGTGCTGACATGGGCTTAGGAGTTCCATTTAATATAGCTAGTTATTCGCTGCTCACACACATGATTGCTCATGTAACTGGTTACCAAGCTGGAGAATTTATACACACAACAGGAGATACCCACGTGTATCTAAATCATATTGAACCATTAAAGATACAGCTACAGAGAGAACCAAAACAGTTTCCAACATTAGAATTTGCTAGAAAAATTGAATCCATTGATGATTTCAAATATGAGGACTTCATTATTAAAGATTACAAACCACATCCTAAAATTGAAATGGAAATGGCAGTATAG
Protein
MTHNNDVEIKSHDEYQYLKLIDQIIKSGDKRVDRTGVGTLSIFGAMQRYSLHNNTLPLLTTKRVFTRGVIEELLWMISGSTDSKALAAKGVHIWDANGSRNFLDNLGFTDREEGDLGPVYGFQWRHSGAKYIDCQTDYTGQGIDQLQSVIDSIKNNPADRRMLICAWNSSDLKKMALPPCHCLAQFYVADGKLSCLLYQRSADMGLGVPFNIASYSLLTHMIAHVTGYQAGEFIHTTGDTHVYLNHIEPLKIQLQREPKQFPTLEFARKIESIDDFKYEDFIIKDYKPHPKIEMEMAV
Summary
Description
Contributes to the de novo mitochondrial thymidylate biosynthesis pathway.
Catalytic Activity
(6R)-5,10-methylene-5,6,7,8-tetrahydrofolate + dUMP = 7,8-dihydrofolate + dTMP
Subunit
Homodimer.
Similarity
Belongs to the thymidylate synthase family.
Keywords
3D-structure
Complete proteome
Cytoplasm
Isopeptide bond
Membrane
Methyltransferase
Mitochondrion
Mitochondrion inner membrane
Nucleotide biosynthesis
Nucleus
Phosphoprotein
Reference proteome
Transferase
Ubl conjugation
Alternative splicing
Direct protein sequencing
Feature
chain Thymidylate synthase
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
Q2F5K5
A0A2A4J8S8
A0A2H1V417
A0A194R4A9
H9IV57
A0A3S2NJ86
+ More
S4P2I3 A0A212FP22 Q2F5K4 A0A0L7K3P8 Q8JKK4 G9I061 B0WW05 A0A1Q3F1A9 Q16JK1 A0A182G6R8 A0A023EMN0 A0A1Q3F135 R7V2Q2 W5JIC5 A0A0P4WZH3 U5ESY8 Q7QDW7 A0A1B6D8E5 A0A182LNB3 A0A182X845 C6GJB8 A0A182QNV2 Q8VDV6 A0A182HXB7 Q544L2 P07607 A0A182PSB4 A0A2Z6REC7 A0A3Q3AQ82 P45352 L7M0X3 A0A182FGX0 A0A3Q0D2T9 F7G371 C4WVP2 A0A182YIR6 A0A067RHH9 A0A2R5LF67 Q9D0H1 G1KHU7 A0A224Z5N7 H0XYX0 M3Y229 A0A182NDN5 A0A3Q1IQE7 K7EU16 G3IFY1 A0A1Y1NMP9 G3TE40 A0A023GI34 A0A3P9AVQ0 I3JFM1 H2NVZ8 A0A2I4BLY1 A0A3P8N9Y3 A0A372R9Z3 A0A2J7RCQ1 A0A1A8ET85 T1G6T0 A0A2R9BCN0 G2HEY4 A0A2S2QRK0 M3WB14 A0A3Q7QJI9 A0A2Y9EAU2 A0A2K5EZV6 A0A2K6ME29 A0A2K5K9G5 A0A287AFN2 A0A2K6SNU8 F7GTE7 A0A2K6D7G8 A0A2K5X3I1 A0A0D9RYF8 A0A096N9Z2 A0A2K6PR41 F6ZR77 A0A2Y9IRZ6 A0A3Q2VSH6 A0A1U7S9P0 A0A2U3WPZ9 A0A1A8HDL6 A0A2K5S5I4 Q53Y97 P04818 A0A2K6GBJ3 A0A151NEK5 A0A3Q4H909 A0A147AUQ4 A0A2Y9GRT1 U6DSH6 A0A3Q4MU32 A8K9A5 A0A2H8TJS4
S4P2I3 A0A212FP22 Q2F5K4 A0A0L7K3P8 Q8JKK4 G9I061 B0WW05 A0A1Q3F1A9 Q16JK1 A0A182G6R8 A0A023EMN0 A0A1Q3F135 R7V2Q2 W5JIC5 A0A0P4WZH3 U5ESY8 Q7QDW7 A0A1B6D8E5 A0A182LNB3 A0A182X845 C6GJB8 A0A182QNV2 Q8VDV6 A0A182HXB7 Q544L2 P07607 A0A182PSB4 A0A2Z6REC7 A0A3Q3AQ82 P45352 L7M0X3 A0A182FGX0 A0A3Q0D2T9 F7G371 C4WVP2 A0A182YIR6 A0A067RHH9 A0A2R5LF67 Q9D0H1 G1KHU7 A0A224Z5N7 H0XYX0 M3Y229 A0A182NDN5 A0A3Q1IQE7 K7EU16 G3IFY1 A0A1Y1NMP9 G3TE40 A0A023GI34 A0A3P9AVQ0 I3JFM1 H2NVZ8 A0A2I4BLY1 A0A3P8N9Y3 A0A372R9Z3 A0A2J7RCQ1 A0A1A8ET85 T1G6T0 A0A2R9BCN0 G2HEY4 A0A2S2QRK0 M3WB14 A0A3Q7QJI9 A0A2Y9EAU2 A0A2K5EZV6 A0A2K6ME29 A0A2K5K9G5 A0A287AFN2 A0A2K6SNU8 F7GTE7 A0A2K6D7G8 A0A2K5X3I1 A0A0D9RYF8 A0A096N9Z2 A0A2K6PR41 F6ZR77 A0A2Y9IRZ6 A0A3Q2VSH6 A0A1U7S9P0 A0A2U3WPZ9 A0A1A8HDL6 A0A2K5S5I4 Q53Y97 P04818 A0A2K6GBJ3 A0A151NEK5 A0A3Q4H909 A0A147AUQ4 A0A2Y9GRT1 U6DSH6 A0A3Q4MU32 A8K9A5 A0A2H8TJS4
EC Number
2.1.1.45
Pubmed
26354079
19121390
23622113
22118469
26227816
12186886
+ More
22355451 17510324 26483478 24945155 23254933 20920257 23761445 12364791 14747013 17210077 20966253 19559812 15489334 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 3444407 3782103 2915925 7711067 9894005 25576852 26319212 17495919 25244985 24845553 28797301 21804562 23929341 29704459 28004739 25186727 22722832 16136131 21484476 17975172 25243066 25362486 22002653 25319552 11181995 2987839 2243092 12706868 16177791 2532645 3839505 2656695 19690332 21269460 21876188 22814378 23186163 28112733 8845352 11329255 11278511 22293439
22355451 17510324 26483478 24945155 23254933 20920257 23761445 12364791 14747013 17210077 20966253 19559812 15489334 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 3444407 3782103 2915925 7711067 9894005 25576852 26319212 17495919 25244985 24845553 28797301 21804562 23929341 29704459 28004739 25186727 22722832 16136131 21484476 17975172 25243066 25362486 22002653 25319552 11181995 2987839 2243092 12706868 16177791 2532645 3839505 2656695 19690332 21269460 21876188 22814378 23186163 28112733 8845352 11329255 11278511 22293439
EMBL
DQ311418
ABD36362.1
NWSH01002453
PCG68269.1
ODYU01000577
SOQ35588.1
+ More
KQ460761 KPJ12517.1 BABH01000553 RSAL01000074 RVE48957.1 GAIX01012000 JAA80560.1 AGBW02004200 OWR55463.1 DQ311419 ABD36363.1 JTDY01011548 KOB55697.1 AF451898 AAM22072.1 JN418988 AEW69584.1 DS232135 EDS35837.1 GFDL01013777 JAV21268.1 CH478005 EAT34454.1 JXUM01148754 JXUM01148755 KQ570629 KXJ68326.1 GAPW01002995 JAC10603.1 GFDL01013783 JAV21262.1 AMQN01005289 AMQN01005290 KB295624 ELU12824.1 ADMH02001155 ETN63871.1 GDRN01043065 JAI67003.1 GANO01003021 JAB56850.1 AAAB01008849 EAA07160.3 GEDC01015339 JAS21959.1 FJ972199 ACS44779.1 AXCN02001746 BC020139 AAH20139.1 APCN01005173 AK035663 CH466524 BAC29144.1 EDL37260.1 M13019 M13352 J02617 M13347 M13348 M13349 M13350 M13351 X14489 BEXD01003190 GBC00474.1 L12138 BC126093 GACK01008251 JAA56783.1 ABLF02032795 AK341607 BAH71962.1 KK852506 KDR22498.1 GGLE01004044 MBY08170.1 AK011435 BAB27620.1 GFPF01012353 MAA23499.1 AAQR03029650 AEYP01011728 AEYP01011729 ABGA01080913 ABGA01080914 ABGA01080915 ABGA01080916 NDHI03003366 PNJ81170.1 JH002476 KE669711 RAZU01000086 EGW14385.1 ERE82571.1 RLQ74643.1 GEZM01002485 JAV97227.1 GBBM01001857 JAC33561.1 AERX01002937 QKKE01000116 RGB37605.1 NEVH01005885 PNF38599.1 HAEB01003174 SBQ49701.1 AMQM01007119 KB097563 ESN94829.1 AJFE02052166 AJFE02052167 AACZ04065249 AK305298 GABC01000484 GABF01007534 GABD01006261 GABE01000097 NBAG03000249 BAK62292.1 JAA10854.1 JAA14611.1 JAA26839.1 JAA44642.1 PNI61355.1 GGMS01011131 MBY80334.1 AANG04003548 AEMK02000045 GAMT01007895 GAMR01005325 GAMQ01006007 JAB03966.1 JAB28607.1 JAB35844.1 AQIA01032860 AQIB01129191 AHZZ02012367 JU322646 JU477046 CM001270 AFE66402.1 AFH33850.1 EHH29203.1 HAEC01014079 SBQ82296.1 BT006811 AK314140 CH471113 AAP35457.1 BAG36830.1 EAX01721.1 X02308 D00596 AB077207 AB077208 AB062290 AP001178 BC002567 BC013919 BC083512 D00517 AKHW03003201 KYO35237.1 GCES01004166 JAR82157.1 HAAF01013649 CCP85471.1 AK292620 BAF85309.1 GFXV01001723 MBW13528.1
KQ460761 KPJ12517.1 BABH01000553 RSAL01000074 RVE48957.1 GAIX01012000 JAA80560.1 AGBW02004200 OWR55463.1 DQ311419 ABD36363.1 JTDY01011548 KOB55697.1 AF451898 AAM22072.1 JN418988 AEW69584.1 DS232135 EDS35837.1 GFDL01013777 JAV21268.1 CH478005 EAT34454.1 JXUM01148754 JXUM01148755 KQ570629 KXJ68326.1 GAPW01002995 JAC10603.1 GFDL01013783 JAV21262.1 AMQN01005289 AMQN01005290 KB295624 ELU12824.1 ADMH02001155 ETN63871.1 GDRN01043065 JAI67003.1 GANO01003021 JAB56850.1 AAAB01008849 EAA07160.3 GEDC01015339 JAS21959.1 FJ972199 ACS44779.1 AXCN02001746 BC020139 AAH20139.1 APCN01005173 AK035663 CH466524 BAC29144.1 EDL37260.1 M13019 M13352 J02617 M13347 M13348 M13349 M13350 M13351 X14489 BEXD01003190 GBC00474.1 L12138 BC126093 GACK01008251 JAA56783.1 ABLF02032795 AK341607 BAH71962.1 KK852506 KDR22498.1 GGLE01004044 MBY08170.1 AK011435 BAB27620.1 GFPF01012353 MAA23499.1 AAQR03029650 AEYP01011728 AEYP01011729 ABGA01080913 ABGA01080914 ABGA01080915 ABGA01080916 NDHI03003366 PNJ81170.1 JH002476 KE669711 RAZU01000086 EGW14385.1 ERE82571.1 RLQ74643.1 GEZM01002485 JAV97227.1 GBBM01001857 JAC33561.1 AERX01002937 QKKE01000116 RGB37605.1 NEVH01005885 PNF38599.1 HAEB01003174 SBQ49701.1 AMQM01007119 KB097563 ESN94829.1 AJFE02052166 AJFE02052167 AACZ04065249 AK305298 GABC01000484 GABF01007534 GABD01006261 GABE01000097 NBAG03000249 BAK62292.1 JAA10854.1 JAA14611.1 JAA26839.1 JAA44642.1 PNI61355.1 GGMS01011131 MBY80334.1 AANG04003548 AEMK02000045 GAMT01007895 GAMR01005325 GAMQ01006007 JAB03966.1 JAB28607.1 JAB35844.1 AQIA01032860 AQIB01129191 AHZZ02012367 JU322646 JU477046 CM001270 AFE66402.1 AFH33850.1 EHH29203.1 HAEC01014079 SBQ82296.1 BT006811 AK314140 CH471113 AAP35457.1 BAG36830.1 EAX01721.1 X02308 D00596 AB077207 AB077208 AB062290 AP001178 BC002567 BC013919 BC083512 D00517 AKHW03003201 KYO35237.1 GCES01004166 JAR82157.1 HAAF01013649 CCP85471.1 AK292620 BAF85309.1 GFXV01001723 MBW13528.1
Proteomes
UP000218220
UP000053240
UP000005204
UP000283053
UP000007151
UP000037510
+ More
UP000232784 UP000029779 UP000002320 UP000008820 UP000069940 UP000249989 UP000014760 UP000000673 UP000007062 UP000075882 UP000076407 UP000075886 UP000075840 UP000000589 UP000075885 UP000247702 UP000264800 UP000002494 UP000069272 UP000189706 UP000002280 UP000007819 UP000076408 UP000027135 UP000001646 UP000005225 UP000000715 UP000075884 UP000265040 UP000001595 UP000001075 UP000030759 UP000273346 UP000007646 UP000265160 UP000005207 UP000192220 UP000265100 UP000263633 UP000235965 UP000015101 UP000240080 UP000002277 UP000011712 UP000286641 UP000248480 UP000233020 UP000233180 UP000233080 UP000008227 UP000233220 UP000008225 UP000233120 UP000233100 UP000029965 UP000028761 UP000233200 UP000248482 UP000264840 UP000189705 UP000245340 UP000233040 UP000005640 UP000233160 UP000050525 UP000261580 UP000265000 UP000248481
UP000232784 UP000029779 UP000002320 UP000008820 UP000069940 UP000249989 UP000014760 UP000000673 UP000007062 UP000075882 UP000076407 UP000075886 UP000075840 UP000000589 UP000075885 UP000247702 UP000264800 UP000002494 UP000069272 UP000189706 UP000002280 UP000007819 UP000076408 UP000027135 UP000001646 UP000005225 UP000000715 UP000075884 UP000265040 UP000001595 UP000001075 UP000030759 UP000273346 UP000007646 UP000265160 UP000005207 UP000192220 UP000265100 UP000263633 UP000235965 UP000015101 UP000240080 UP000002277 UP000011712 UP000286641 UP000248480 UP000233020 UP000233180 UP000233080 UP000008227 UP000233220 UP000008225 UP000233120 UP000233100 UP000029965 UP000028761 UP000233200 UP000248482 UP000264840 UP000189705 UP000245340 UP000233040 UP000005640 UP000233160 UP000050525 UP000261580 UP000265000 UP000248481
Pfam
PF00303 Thymidylat_synt
Interpro
SUPFAM
SSF55831
SSF55831
Gene 3D
ProteinModelPortal
Q2F5K5
A0A2A4J8S8
A0A2H1V417
A0A194R4A9
H9IV57
A0A3S2NJ86
+ More
S4P2I3 A0A212FP22 Q2F5K4 A0A0L7K3P8 Q8JKK4 G9I061 B0WW05 A0A1Q3F1A9 Q16JK1 A0A182G6R8 A0A023EMN0 A0A1Q3F135 R7V2Q2 W5JIC5 A0A0P4WZH3 U5ESY8 Q7QDW7 A0A1B6D8E5 A0A182LNB3 A0A182X845 C6GJB8 A0A182QNV2 Q8VDV6 A0A182HXB7 Q544L2 P07607 A0A182PSB4 A0A2Z6REC7 A0A3Q3AQ82 P45352 L7M0X3 A0A182FGX0 A0A3Q0D2T9 F7G371 C4WVP2 A0A182YIR6 A0A067RHH9 A0A2R5LF67 Q9D0H1 G1KHU7 A0A224Z5N7 H0XYX0 M3Y229 A0A182NDN5 A0A3Q1IQE7 K7EU16 G3IFY1 A0A1Y1NMP9 G3TE40 A0A023GI34 A0A3P9AVQ0 I3JFM1 H2NVZ8 A0A2I4BLY1 A0A3P8N9Y3 A0A372R9Z3 A0A2J7RCQ1 A0A1A8ET85 T1G6T0 A0A2R9BCN0 G2HEY4 A0A2S2QRK0 M3WB14 A0A3Q7QJI9 A0A2Y9EAU2 A0A2K5EZV6 A0A2K6ME29 A0A2K5K9G5 A0A287AFN2 A0A2K6SNU8 F7GTE7 A0A2K6D7G8 A0A2K5X3I1 A0A0D9RYF8 A0A096N9Z2 A0A2K6PR41 F6ZR77 A0A2Y9IRZ6 A0A3Q2VSH6 A0A1U7S9P0 A0A2U3WPZ9 A0A1A8HDL6 A0A2K5S5I4 Q53Y97 P04818 A0A2K6GBJ3 A0A151NEK5 A0A3Q4H909 A0A147AUQ4 A0A2Y9GRT1 U6DSH6 A0A3Q4MU32 A8K9A5 A0A2H8TJS4
S4P2I3 A0A212FP22 Q2F5K4 A0A0L7K3P8 Q8JKK4 G9I061 B0WW05 A0A1Q3F1A9 Q16JK1 A0A182G6R8 A0A023EMN0 A0A1Q3F135 R7V2Q2 W5JIC5 A0A0P4WZH3 U5ESY8 Q7QDW7 A0A1B6D8E5 A0A182LNB3 A0A182X845 C6GJB8 A0A182QNV2 Q8VDV6 A0A182HXB7 Q544L2 P07607 A0A182PSB4 A0A2Z6REC7 A0A3Q3AQ82 P45352 L7M0X3 A0A182FGX0 A0A3Q0D2T9 F7G371 C4WVP2 A0A182YIR6 A0A067RHH9 A0A2R5LF67 Q9D0H1 G1KHU7 A0A224Z5N7 H0XYX0 M3Y229 A0A182NDN5 A0A3Q1IQE7 K7EU16 G3IFY1 A0A1Y1NMP9 G3TE40 A0A023GI34 A0A3P9AVQ0 I3JFM1 H2NVZ8 A0A2I4BLY1 A0A3P8N9Y3 A0A372R9Z3 A0A2J7RCQ1 A0A1A8ET85 T1G6T0 A0A2R9BCN0 G2HEY4 A0A2S2QRK0 M3WB14 A0A3Q7QJI9 A0A2Y9EAU2 A0A2K5EZV6 A0A2K6ME29 A0A2K5K9G5 A0A287AFN2 A0A2K6SNU8 F7GTE7 A0A2K6D7G8 A0A2K5X3I1 A0A0D9RYF8 A0A096N9Z2 A0A2K6PR41 F6ZR77 A0A2Y9IRZ6 A0A3Q2VSH6 A0A1U7S9P0 A0A2U3WPZ9 A0A1A8HDL6 A0A2K5S5I4 Q53Y97 P04818 A0A2K6GBJ3 A0A151NEK5 A0A3Q4H909 A0A147AUQ4 A0A2Y9GRT1 U6DSH6 A0A3Q4MU32 A8K9A5 A0A2H8TJS4
PDB
6GKO
E-value=4.41563e-124,
Score=1137
Ontologies
PATHWAY
GO
GO:0004799
GO:0006231
GO:0005739
GO:0005829
GO:0051216
GO:0097421
GO:0051384
GO:0048589
GO:0032570
GO:0007568
GO:0048037
GO:0019860
GO:0035999
GO:0005542
GO:0042803
GO:0060574
GO:0033189
GO:0045471
GO:0046078
GO:0046683
GO:0005759
GO:0007623
GO:0005743
GO:0000166
GO:0034097
GO:1990825
GO:0051593
GO:0000900
GO:0005730
GO:0019088
GO:0005634
GO:0005737
GO:0017148
GO:0006235
GO:0003729
GO:0006417
GO:0008144
GO:0042493
GO:0009636
GO:0014070
GO:0046653
GO:0006206
GO:0004146
GO:0006730
GO:0060041
GO:0015949
GO:0071897
GO:0000083
GO:0006468
GO:0005509
GO:0016020
GO:0005524
GO:0006412
GO:0016876
GO:0043039
Topology
Subcellular location
Nucleus
Cytoplasm
Mitochondrion
Mitochondrion matrix
Mitochondrion inner membrane
Cytoplasm
Mitochondrion
Mitochondrion matrix
Mitochondrion inner membrane
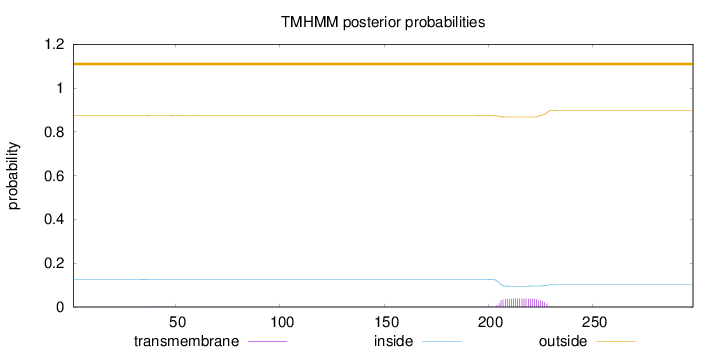
Length:
298
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.85494
Exp number, first 60 AAs:
0.02412
Total prob of N-in:
0.12680
outside
1 - 298
Population Genetic Test Statistics
Pi
210.022803
Theta
181.793556
Tajima's D
0.509365
CLR
38.741305
CSRT
0.521673916304185
Interpretation
Uncertain
