Pre Gene Modal
BGIBMGA001138
Annotation
PREDICTED:_putative_neutral_sphingomyelinase_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.363
Sequence
CDS
ATGGGTTATAGAGTATTTACTGAGCATTTAACATTTAATATACATCTTAAATTTTATTTAAAAACAAGTCGTCATCTAACTAAATTAAGTTTCTTTAGAAACCACATATACATACTTCAAAGGGGTATACCAGTGGTGTCTAAGAATAAGAAAGAAAGATATGAAGCTATATCTACATACTTATTAAAAAGCGAACATAATATAGTATGTTTGCAAGAAGTTTGGAGTGAAAAAGATTACCTATATTTAAAGGAGAACCTAAAGAATGTACTCCCATATTCTTACTACTTTTACAGTGGAGTTCTAGGTTCAGGTCTCTGTGTATTTTCTAAATGGGTAATCCAAGATGTCTTCTTCCATCAGTGGCCCTTGAATGGGTACATACACAAAATACATCATGGTGACTGGTTTGGAGGAAAAGGTGTGGGTCTCTGTCGGATCAAGTTTGGGGAGAGACTCATCAATGTCTATTGCACACATCTTCACGCAGAATACCACGAAGACGACATGTATCTAGCTCATCGTGTGCTTCAGGCATACTCAACAGCAGAGTTCGTCAAATTAACATCATCACCAGCTGATGTTTCTATACTAGCTGGAGATTTAAATACTGCACCCGGGGATTTGTCTTATAAGATAATATCCCAACTGCCATCTCTTCTTGATCCATATAACATGAAGTTTGAAGGGACCAATCCTTTAATAGCTAAAGCATCTGGTACAAGTGATAACCTCAACAATAGCTATTCGGATCCTAAACAAGTAAAGGCATATCCTGAAGGGAAACGTATAGATCATATATTATTCCATACTAACCATTCTTGGGAGGCCCGTGTCGTGAATTTCGGTAATCCGTTACCAGATAGAGTTCCAGAACAACAGTTTTCTTACTCTGATCACAATGCAGTCTCGCTTGAGTTACACCTAAAACCTTGTGAACAAAAATTAAACCAGAGACAAGAGAGTGTAGACAGTGCATTTCAAGAAACTATCACTCAAGCGATCAAAGTTTGCAGGGATGCCACAACAAATTTATCGAAATCAAAAAAGCTGTATTTACTCAGCGGCGGTTTAATATTCATGTTTTTGTTAGGTTCAGTAGGTTTCTGGCCTAACTTTCTTATTTATGATGTACTGAAACTTCTTATTACAGCCTTATGCTTCTACAATTTAGTAATGGGATCTCTATGGAATCAAATTGAAATGAACAGTTTGAAAGCTGGTCTGAATGCCTTAGAGAATTTCATTCAAACAAGAAATGATACAAAACCCGAAGACATCAATAAAGAGGATGTTAAAAACAAATGA
Protein
MGYRVFTEHLTFNIHLKFYLKTSRHLTKLSFFRNHIYILQRGIPVVSKNKKERYEAISTYLLKSEHNIVCLQEVWSEKDYLYLKENLKNVLPYSYYFYSGVLGSGLCVFSKWVIQDVFFHQWPLNGYIHKIHHGDWFGGKGVGLCRIKFGERLINVYCTHLHAEYHEDDMYLAHRVLQAYSTAEFVKLTSSPADVSILAGDLNTAPGDLSYKIISQLPSLLDPYNMKFEGTNPLIAKASGTSDNLNNSYSDPKQVKAYPEGKRIDHILFHTNHSWEARVVNFGNPLPDRVPEQQFSYSDHNAVSLELHLKPCEQKLNQRQESVDSAFQETITQAIKVCRDATTNLSKSKKLYLLSGGLIFMFLLGSVGFWPNFLIYDVLKLLITALCFYNLVMGSLWNQIEMNSLKAGLNALENFIQTRNDTKPEDINKEDVKNK
Summary
Uniprot
H9IV58
A0A194R4T0
A0A194Q7I2
A0A2A4JA21
A0A1E1WTB4
S4P819
+ More
A0A0L7LAW8 A0A195BUS2 A0A158NEL9 F4WRU1 A0A195CQW9 A0A195FMQ1 A0A195EMZ9 E9IJD8 A0A026WUF7 E2AEJ5 A0A151XJ16 A0A067QJ62 K7J2F3 A0A154PE23 A0A232EY38 D1ZZU9 A0A087ZRR4 A0A0M9A5E9 A0A2A3EQG7 A0A0L7R5L5 A0A1W4X3U6 A0A0C9QFV7 U4U6L4 N6U1V5 A0A2U6TC85 A0A182Y030 A0A182LZ60 B0W7J5 A7UUQ6 A0A182NU79 Q17JK7 A0A182TGU1 A0A1S4GXT5 A0A182HUV9 A0A182L1P3 B0XLI7 A0A182V8B4 A0A182GNG4 A0A2M4AND1 A0A182WU92 A0A182P4W5 A0A182RC91 A0A182QW34 A0A182W797 A0A336KTK6 A0A182GXD5 A0A226E771 A0A0K8TSH4 A0A336KQA9 W5JLV8 A0A182KDE3 A0A1B6DDU6 A0A1B6E4I2 A0A1B6IHQ4 A0A1Q3FAF3 A0A182J841 A0A182F6D9 A0A310SHV1 U5EWI2 A0A1Y1K207 A0A084VNC4 A0A2J7RG53 A0A0P5KZZ5 A0A0P5G7K4 A0A1D2MWX7 A0A0P5PDD6 A0A0P5E581 A0A0P6JEK1 A0A1L8DJX7 A0A0P5PP10 E9GIK1 A0A1L8DJZ2 A0A0P5HVW5 A0A0K8T9T5 A0A1J1HZR6 A0A1B0DRC6 A0A0A9XVL4 A0A0P5KGP8 A0A0M4EC29 A0A0P5LTP6 A0A0L0CEY0 A0A0Q9XLV1 A0A1D2N153 A0A0A1WKI1 W8BLR8 A0A069DZ32
A0A0L7LAW8 A0A195BUS2 A0A158NEL9 F4WRU1 A0A195CQW9 A0A195FMQ1 A0A195EMZ9 E9IJD8 A0A026WUF7 E2AEJ5 A0A151XJ16 A0A067QJ62 K7J2F3 A0A154PE23 A0A232EY38 D1ZZU9 A0A087ZRR4 A0A0M9A5E9 A0A2A3EQG7 A0A0L7R5L5 A0A1W4X3U6 A0A0C9QFV7 U4U6L4 N6U1V5 A0A2U6TC85 A0A182Y030 A0A182LZ60 B0W7J5 A7UUQ6 A0A182NU79 Q17JK7 A0A182TGU1 A0A1S4GXT5 A0A182HUV9 A0A182L1P3 B0XLI7 A0A182V8B4 A0A182GNG4 A0A2M4AND1 A0A182WU92 A0A182P4W5 A0A182RC91 A0A182QW34 A0A182W797 A0A336KTK6 A0A182GXD5 A0A226E771 A0A0K8TSH4 A0A336KQA9 W5JLV8 A0A182KDE3 A0A1B6DDU6 A0A1B6E4I2 A0A1B6IHQ4 A0A1Q3FAF3 A0A182J841 A0A182F6D9 A0A310SHV1 U5EWI2 A0A1Y1K207 A0A084VNC4 A0A2J7RG53 A0A0P5KZZ5 A0A0P5G7K4 A0A1D2MWX7 A0A0P5PDD6 A0A0P5E581 A0A0P6JEK1 A0A1L8DJX7 A0A0P5PP10 E9GIK1 A0A1L8DJZ2 A0A0P5HVW5 A0A0K8T9T5 A0A1J1HZR6 A0A1B0DRC6 A0A0A9XVL4 A0A0P5KGP8 A0A0M4EC29 A0A0P5LTP6 A0A0L0CEY0 A0A0Q9XLV1 A0A1D2N153 A0A0A1WKI1 W8BLR8 A0A069DZ32
Pubmed
19121390
26354079
23622113
26227816
21347285
21719571
+ More
21282665 24508170 30249741 20798317 24845553 20075255 28648823 18362917 19820115 23537049 25244985 12364791 17510324 20966253 26483478 26369729 20920257 23761445 28004739 24438588 27289101 21292972 25401762 26823975 26108605 17994087 25830018 24495485 26334808
21282665 24508170 30249741 20798317 24845553 20075255 28648823 18362917 19820115 23537049 25244985 12364791 17510324 20966253 26483478 26369729 20920257 23761445 28004739 24438588 27289101 21292972 25401762 26823975 26108605 17994087 25830018 24495485 26334808
EMBL
BABH01000557
KQ460761
KPJ12519.1
KQ459439
KPJ00955.1
NWSH01002453
+ More
PCG68272.1 GDQN01000877 JAT90177.1 GAIX01004344 JAA88216.1 JTDY01001914 KOB72550.1 KQ976401 KYM92369.1 ADTU01013497 GL888292 EGI63098.1 KQ977381 KYN03101.1 KQ981490 KYN41214.1 KQ978625 KYN29578.1 GL763764 EFZ19335.1 KK107119 QOIP01000009 EZA58744.1 RLU18828.1 GL438862 EFN68152.1 KQ982080 KYQ60305.1 KK853449 KDR07562.1 KQ434886 KZC10093.1 NNAY01001687 OXU23225.1 KQ971338 EFA01795.1 KQ435740 KOX76826.1 KZ288202 PBC33512.1 KQ414648 KOC66175.1 GBYB01013403 JAG83170.1 KB631724 ERL85595.1 APGK01045762 APGK01045763 KB741038 ENN74581.1 KX013771 AND65757.1 AXCM01004940 DS231854 EDS38030.1 AAAB01008964 EDO63591.1 CH477232 EAT46833.1 APCN01002480 DS234497 EDS34220.1 JXUM01076253 KQ562934 KXJ74809.1 GGFK01008970 MBW42291.1 AXCN02000772 UFQS01000880 UFQT01000880 SSX07472.1 SSX27812.1 JXUM01021513 KQ560603 KXJ81668.1 LNIX01000006 OXA52817.1 GDAI01000718 JAI16885.1 UFQS01000798 UFQT01000798 SSX06986.1 SSX27330.1 ADMH02001153 ETN63880.1 GEDC01013421 JAS23877.1 GEDC01004509 JAS32789.1 GECU01021257 JAS86449.1 GFDL01010481 JAV24564.1 KQ759874 OAD62358.1 GANO01001404 JAB58467.1 GEZM01096908 GEZM01096903 GEZM01096896 JAV54488.1 ATLV01014737 KE524984 KFB39468.1 NEVH01004406 PNF39814.1 GDIQ01202513 GDIQ01202512 JAK49213.1 GDIQ01245157 GDIQ01222308 GDIQ01222307 GDIQ01164188 GDIQ01158177 GDIQ01149133 GDIQ01137433 GDIQ01125701 GDIQ01116549 GDIQ01109050 GDIQ01076353 GDIQ01071310 JAK06568.1 LJIJ01000465 ODM97185.1 GDIQ01130240 JAL21486.1 GDIP01147901 LRGB01000568 JAJ75501.1 KZS18109.1 GDIQ01008621 JAN86116.1 GFDF01007338 JAV06746.1 GDIQ01131497 JAL20229.1 GL732546 EFX80740.1 GFDF01007318 JAV06766.1 GDIQ01227111 JAK24614.1 GBRD01003514 JAG62307.1 CVRI01000037 CRK93599.1 AJVK01019877 GBHO01020179 GBHO01020178 GDHC01008207 JAG23425.1 JAG23426.1 JAQ10422.1 GDIQ01191158 JAK60567.1 CP012525 ALC42908.1 GDIQ01165780 JAK85945.1 JRES01000493 KNC30805.1 CH933809 KRG06130.1 LJIJ01000303 ODM99023.1 GBXI01015252 GBXI01011982 JAC99039.1 JAD02310.1 GAMC01006793 JAB99762.1 GBGD01001345 JAC87544.1
PCG68272.1 GDQN01000877 JAT90177.1 GAIX01004344 JAA88216.1 JTDY01001914 KOB72550.1 KQ976401 KYM92369.1 ADTU01013497 GL888292 EGI63098.1 KQ977381 KYN03101.1 KQ981490 KYN41214.1 KQ978625 KYN29578.1 GL763764 EFZ19335.1 KK107119 QOIP01000009 EZA58744.1 RLU18828.1 GL438862 EFN68152.1 KQ982080 KYQ60305.1 KK853449 KDR07562.1 KQ434886 KZC10093.1 NNAY01001687 OXU23225.1 KQ971338 EFA01795.1 KQ435740 KOX76826.1 KZ288202 PBC33512.1 KQ414648 KOC66175.1 GBYB01013403 JAG83170.1 KB631724 ERL85595.1 APGK01045762 APGK01045763 KB741038 ENN74581.1 KX013771 AND65757.1 AXCM01004940 DS231854 EDS38030.1 AAAB01008964 EDO63591.1 CH477232 EAT46833.1 APCN01002480 DS234497 EDS34220.1 JXUM01076253 KQ562934 KXJ74809.1 GGFK01008970 MBW42291.1 AXCN02000772 UFQS01000880 UFQT01000880 SSX07472.1 SSX27812.1 JXUM01021513 KQ560603 KXJ81668.1 LNIX01000006 OXA52817.1 GDAI01000718 JAI16885.1 UFQS01000798 UFQT01000798 SSX06986.1 SSX27330.1 ADMH02001153 ETN63880.1 GEDC01013421 JAS23877.1 GEDC01004509 JAS32789.1 GECU01021257 JAS86449.1 GFDL01010481 JAV24564.1 KQ759874 OAD62358.1 GANO01001404 JAB58467.1 GEZM01096908 GEZM01096903 GEZM01096896 JAV54488.1 ATLV01014737 KE524984 KFB39468.1 NEVH01004406 PNF39814.1 GDIQ01202513 GDIQ01202512 JAK49213.1 GDIQ01245157 GDIQ01222308 GDIQ01222307 GDIQ01164188 GDIQ01158177 GDIQ01149133 GDIQ01137433 GDIQ01125701 GDIQ01116549 GDIQ01109050 GDIQ01076353 GDIQ01071310 JAK06568.1 LJIJ01000465 ODM97185.1 GDIQ01130240 JAL21486.1 GDIP01147901 LRGB01000568 JAJ75501.1 KZS18109.1 GDIQ01008621 JAN86116.1 GFDF01007338 JAV06746.1 GDIQ01131497 JAL20229.1 GL732546 EFX80740.1 GFDF01007318 JAV06766.1 GDIQ01227111 JAK24614.1 GBRD01003514 JAG62307.1 CVRI01000037 CRK93599.1 AJVK01019877 GBHO01020179 GBHO01020178 GDHC01008207 JAG23425.1 JAG23426.1 JAQ10422.1 GDIQ01191158 JAK60567.1 CP012525 ALC42908.1 GDIQ01165780 JAK85945.1 JRES01000493 KNC30805.1 CH933809 KRG06130.1 LJIJ01000303 ODM99023.1 GBXI01015252 GBXI01011982 JAC99039.1 JAD02310.1 GAMC01006793 JAB99762.1 GBGD01001345 JAC87544.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000078540
+ More
UP000005205 UP000007755 UP000078542 UP000078541 UP000078492 UP000053097 UP000279307 UP000000311 UP000075809 UP000027135 UP000002358 UP000076502 UP000215335 UP000007266 UP000005203 UP000053105 UP000242457 UP000053825 UP000192223 UP000030742 UP000019118 UP000076408 UP000075883 UP000002320 UP000007062 UP000075884 UP000008820 UP000075902 UP000075840 UP000075882 UP000075903 UP000069940 UP000249989 UP000076407 UP000075885 UP000075900 UP000075886 UP000075920 UP000198287 UP000000673 UP000075881 UP000075880 UP000069272 UP000030765 UP000235965 UP000094527 UP000076858 UP000000305 UP000183832 UP000092462 UP000092553 UP000037069 UP000009192
UP000005205 UP000007755 UP000078542 UP000078541 UP000078492 UP000053097 UP000279307 UP000000311 UP000075809 UP000027135 UP000002358 UP000076502 UP000215335 UP000007266 UP000005203 UP000053105 UP000242457 UP000053825 UP000192223 UP000030742 UP000019118 UP000076408 UP000075883 UP000002320 UP000007062 UP000075884 UP000008820 UP000075902 UP000075840 UP000075882 UP000075903 UP000069940 UP000249989 UP000076407 UP000075885 UP000075900 UP000075886 UP000075920 UP000198287 UP000000673 UP000075881 UP000075880 UP000069272 UP000030765 UP000235965 UP000094527 UP000076858 UP000000305 UP000183832 UP000092462 UP000092553 UP000037069 UP000009192
PRIDE
Interpro
IPR036691
Endo/exonu/phosph_ase_sf
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR038772 SMPD2-like
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR036682 OS_D_A10/PebIII_sf
IPR005055 A10/PebIII
IPR034016 M1_APN-typ
IPR001930 Peptidase_M1
IPR014782 Peptidase_M1_dom
IPR024571 ERAP1-like_C_dom
IPR042097 Aminopeptidase_N-like_N
IPR005135 Endo/exonuclease/phosphatase
IPR038772 SMPD2-like
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR036682 OS_D_A10/PebIII_sf
IPR005055 A10/PebIII
IPR034016 M1_APN-typ
IPR001930 Peptidase_M1
IPR014782 Peptidase_M1_dom
IPR024571 ERAP1-like_C_dom
IPR042097 Aminopeptidase_N-like_N
Gene 3D
CDD
ProteinModelPortal
H9IV58
A0A194R4T0
A0A194Q7I2
A0A2A4JA21
A0A1E1WTB4
S4P819
+ More
A0A0L7LAW8 A0A195BUS2 A0A158NEL9 F4WRU1 A0A195CQW9 A0A195FMQ1 A0A195EMZ9 E9IJD8 A0A026WUF7 E2AEJ5 A0A151XJ16 A0A067QJ62 K7J2F3 A0A154PE23 A0A232EY38 D1ZZU9 A0A087ZRR4 A0A0M9A5E9 A0A2A3EQG7 A0A0L7R5L5 A0A1W4X3U6 A0A0C9QFV7 U4U6L4 N6U1V5 A0A2U6TC85 A0A182Y030 A0A182LZ60 B0W7J5 A7UUQ6 A0A182NU79 Q17JK7 A0A182TGU1 A0A1S4GXT5 A0A182HUV9 A0A182L1P3 B0XLI7 A0A182V8B4 A0A182GNG4 A0A2M4AND1 A0A182WU92 A0A182P4W5 A0A182RC91 A0A182QW34 A0A182W797 A0A336KTK6 A0A182GXD5 A0A226E771 A0A0K8TSH4 A0A336KQA9 W5JLV8 A0A182KDE3 A0A1B6DDU6 A0A1B6E4I2 A0A1B6IHQ4 A0A1Q3FAF3 A0A182J841 A0A182F6D9 A0A310SHV1 U5EWI2 A0A1Y1K207 A0A084VNC4 A0A2J7RG53 A0A0P5KZZ5 A0A0P5G7K4 A0A1D2MWX7 A0A0P5PDD6 A0A0P5E581 A0A0P6JEK1 A0A1L8DJX7 A0A0P5PP10 E9GIK1 A0A1L8DJZ2 A0A0P5HVW5 A0A0K8T9T5 A0A1J1HZR6 A0A1B0DRC6 A0A0A9XVL4 A0A0P5KGP8 A0A0M4EC29 A0A0P5LTP6 A0A0L0CEY0 A0A0Q9XLV1 A0A1D2N153 A0A0A1WKI1 W8BLR8 A0A069DZ32
A0A0L7LAW8 A0A195BUS2 A0A158NEL9 F4WRU1 A0A195CQW9 A0A195FMQ1 A0A195EMZ9 E9IJD8 A0A026WUF7 E2AEJ5 A0A151XJ16 A0A067QJ62 K7J2F3 A0A154PE23 A0A232EY38 D1ZZU9 A0A087ZRR4 A0A0M9A5E9 A0A2A3EQG7 A0A0L7R5L5 A0A1W4X3U6 A0A0C9QFV7 U4U6L4 N6U1V5 A0A2U6TC85 A0A182Y030 A0A182LZ60 B0W7J5 A7UUQ6 A0A182NU79 Q17JK7 A0A182TGU1 A0A1S4GXT5 A0A182HUV9 A0A182L1P3 B0XLI7 A0A182V8B4 A0A182GNG4 A0A2M4AND1 A0A182WU92 A0A182P4W5 A0A182RC91 A0A182QW34 A0A182W797 A0A336KTK6 A0A182GXD5 A0A226E771 A0A0K8TSH4 A0A336KQA9 W5JLV8 A0A182KDE3 A0A1B6DDU6 A0A1B6E4I2 A0A1B6IHQ4 A0A1Q3FAF3 A0A182J841 A0A182F6D9 A0A310SHV1 U5EWI2 A0A1Y1K207 A0A084VNC4 A0A2J7RG53 A0A0P5KZZ5 A0A0P5G7K4 A0A1D2MWX7 A0A0P5PDD6 A0A0P5E581 A0A0P6JEK1 A0A1L8DJX7 A0A0P5PP10 E9GIK1 A0A1L8DJZ2 A0A0P5HVW5 A0A0K8T9T5 A0A1J1HZR6 A0A1B0DRC6 A0A0A9XVL4 A0A0P5KGP8 A0A0M4EC29 A0A0P5LTP6 A0A0L0CEY0 A0A0Q9XLV1 A0A1D2N153 A0A0A1WKI1 W8BLR8 A0A069DZ32
PDB
5UVG
E-value=9.0922e-05,
Score=109
Ontologies
PATHWAY
GO
PANTHER
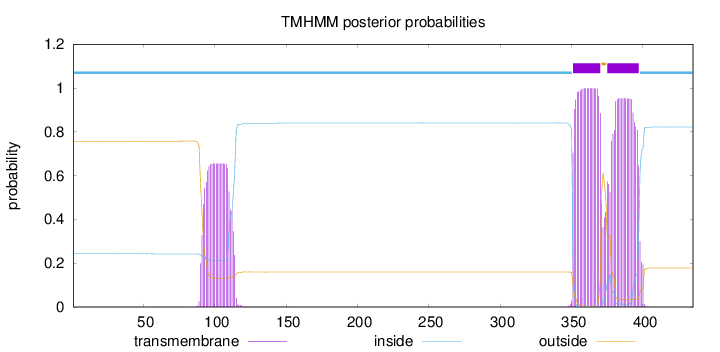
Topology
Length:
435
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
55.22693
Exp number, first 60 AAs:
0.02916
Total prob of N-in:
0.24366
inside
1 - 350
TMhelix
351 - 370
outside
371 - 374
TMhelix
375 - 397
inside
398 - 435
Population Genetic Test Statistics
Pi
231.311543
Theta
195.06987
Tajima's D
0.844802
CLR
0.308066
CSRT
0.614569271536423
Interpretation
Uncertain
