Gene
KWMTBOMO07999
Pre Gene Modal
BGIBMGA001013
Annotation
putative_cAMP-dependent_protein_kinase_catalytic_subunit?_partial_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.895
Sequence
CDS
ATGGGGGGCGGCGATAAGATAGACCTTAGCACAACACAATCGGACATAGCCGTTTTACAGGAAAAGGGATTCATACTAGAAAAAGTTATCGGAGAAGGTTCTTACGCTAAGGTTTACAAAGCCACTCATATGGTAGACGAGACCCGGCATTCAATAATGGCCTGTAAAGTCATCGACACAGCTAAGGCGCCCAGGGACTATCTCACAAAGTTCTTGCCTCGCGAGCTAGATGTTCTGATACGCATCAACCACCCTCACATCGTGCACGTGTCCAACATATTTCAACGTCGCGCTAAATACTTTATTTTTTTACGATTTGCTGAAAACGGCGATCTGTTGGACTTTCTCACTCAAAACGGGGCAATACCGGAAAATCAAAGCAGACTGTGGATGCGGCAAATCATATCCGGCCTTCAATACATTCACACAATGAACATAGCTCACAGAGATCTAAAATGCGAAAACGTTTTGGTCACGGCCAATTATAATGTTAAAATAACTGATTTTGGATTTGCACGTAATGTGCGTCAACGAGATAGAGACGTTCTAAGCGAGACGTACTGTGGTTCTCTTTCGTACGCCGCTCCCGAAGTACTAAAGGGGGTGCCTTACATGCCTAAACTGGCCGATATGTGGTCGATCGGCGTTATACTATACACCATGCTCAACAAAGCTTTACCTTTCAACGAAACGTCCGTCAAAAAGCTGTACGAAAAACAGAACTAA
Protein
MGGGDKIDLSTTQSDIAVLQEKGFILEKVIGEGSYAKVYKATHMVDETRHSIMACKVIDTAKAPRDYLTKFLPRELDVLIRINHPHIVHVSNIFQRRAKYFIFLRFAENGDLLDFLTQNGAIPENQSRLWMRQIISGLQYIHTMNIAHRDLKCENVLVTANYNVKITDFGFARNVRQRDRDVLSETYCGSLSYAAPEVLKGVPYMPKLADMWSIGVILYTMLNKALPFNETSVKKLYEKQN
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
H9IUT3
A0A0L7LN37
A0A212FNY0
A0A194Q8K1
A0A2A4J1M8
A0A194R3Y9
+ More
A0A2H1V438 E0VUJ8 A0A026W6M0 E9IFJ5 A0A310SBN0 A0A1W4XJG4 F4X876 A0A087ZQW3 A0A2A3E208 A0A0L7KZB1 A0A151IMD4 E1ZVD3 A0A0C9RFD0 A0A195DR43 A0A151X9F2 A0A0M8ZY63 A0A158NC21 A0A0J7KQI0 A0A195BG99 A0A139WJG5 H9IUT2 A0A212FP11 A0A2H1V427 A0A194R9I7 A0A2A4J2V2 A0A194Q8W4 A0A0L7R0R0 A0A154PAK5 A0A1D2N9J0 A0A336M858 A0A336MBU8 N6UFU9 A0A0L7KQT7 H9J602 A0A194RKW2 A0A194PHV9 A0A2P8XD25 A0A154P4L0 A0A2H1WJA0 A0A2A4K6Z1 A0A0L7R1E4 A0A212ENN5 E2BUI7 A0A2A3ERW8 A0A088AFP6 A0A0M8ZWN2 G8GJE9 A0A1Y1LVT0 D6WR20 N6SSY9 A0A0A9WX60 A0A195DLT1 A0A195FRM3 A0A158NAR6 F4X1E3 E0VNF8 A0A2P8YBE4 A0A026WRP4 A0A1B6IZ62 A0A3Q0IUA1 A0A0C9QW83 A0A1S3D243 A0A195BKA0 A0A1B6FJH7 A0A224XRB7 E9IXN5 A0A1B6MP88 A0A151WWS3 A0A1I8PN93 A0A336MTK5 B4NH25 B4M3X9 B3LW85 A0A0L7LMS7 A0A1I8MVU7 A0A1W4UJJ2 A0A0M3QXR1 A0A0M4EQ10 Q299N1 B4G5G4 A0A3B0K176 B3NZ44 B4JF01 B4K8M4 A0A1A9W2S2 B4PLY8 A0A1B0C7H9 A0A1B0FEQ0 A0A1A9Y9F6 A0A1A9ZE95 Q9VE58 B4QU12 W8BB14 B4I275
A0A2H1V438 E0VUJ8 A0A026W6M0 E9IFJ5 A0A310SBN0 A0A1W4XJG4 F4X876 A0A087ZQW3 A0A2A3E208 A0A0L7KZB1 A0A151IMD4 E1ZVD3 A0A0C9RFD0 A0A195DR43 A0A151X9F2 A0A0M8ZY63 A0A158NC21 A0A0J7KQI0 A0A195BG99 A0A139WJG5 H9IUT2 A0A212FP11 A0A2H1V427 A0A194R9I7 A0A2A4J2V2 A0A194Q8W4 A0A0L7R0R0 A0A154PAK5 A0A1D2N9J0 A0A336M858 A0A336MBU8 N6UFU9 A0A0L7KQT7 H9J602 A0A194RKW2 A0A194PHV9 A0A2P8XD25 A0A154P4L0 A0A2H1WJA0 A0A2A4K6Z1 A0A0L7R1E4 A0A212ENN5 E2BUI7 A0A2A3ERW8 A0A088AFP6 A0A0M8ZWN2 G8GJE9 A0A1Y1LVT0 D6WR20 N6SSY9 A0A0A9WX60 A0A195DLT1 A0A195FRM3 A0A158NAR6 F4X1E3 E0VNF8 A0A2P8YBE4 A0A026WRP4 A0A1B6IZ62 A0A3Q0IUA1 A0A0C9QW83 A0A1S3D243 A0A195BKA0 A0A1B6FJH7 A0A224XRB7 E9IXN5 A0A1B6MP88 A0A151WWS3 A0A1I8PN93 A0A336MTK5 B4NH25 B4M3X9 B3LW85 A0A0L7LMS7 A0A1I8MVU7 A0A1W4UJJ2 A0A0M3QXR1 A0A0M4EQ10 Q299N1 B4G5G4 A0A3B0K176 B3NZ44 B4JF01 B4K8M4 A0A1A9W2S2 B4PLY8 A0A1B0C7H9 A0A1B0FEQ0 A0A1A9Y9F6 A0A1A9ZE95 Q9VE58 B4QU12 W8BB14 B4I275
Pubmed
19121390
26227816
22118469
26354079
20566863
24508170
+ More
30249741 21282665 21719571 20798317 21347285 18362917 19820115 27289101 23537049 29403074 28004739 25401762 17994087 25315136 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485
30249741 21282665 21719571 20798317 21347285 18362917 19820115 27289101 23537049 29403074 28004739 25401762 17994087 25315136 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485
EMBL
BABH01000557
JTDY01000506
KOB76862.1
AGBW02004200
OWR55454.1
KQ459299
+ More
KPJ01853.1 NWSH01003604 PCG66067.1 KQ460761 KPJ12523.1 ODYU01000577 SOQ35597.1 AAZO01005497 DS235786 EEB17054.1 KK107372 QOIP01000004 EZA51705.1 RLU23740.1 GL762841 EFZ20670.1 KQ764887 OAD54258.1 GL888932 EGI57207.1 KZ288439 PBC25730.1 JTDY01004102 KOB68598.1 KQ977059 KYN06039.1 GL434492 EFN74843.1 GBYB01015269 JAG85036.1 KQ980581 KYN15282.1 KQ982373 KYQ56993.1 KQ435826 KOX71989.1 ADTU01011290 LBMM01004298 KMQ92593.1 KQ976500 KYM83202.1 KQ971338 KYB28052.1 OWR55453.1 SOQ35598.1 KPJ12526.1 PCG66066.1 KPJ01854.1 KQ414670 KOC64434.1 KQ434857 KZC08871.1 LJIJ01000133 ODN01930.1 UFQT01000658 SSX26220.1 UFQS01000871 UFQT01000871 SSX07413.1 SSX27755.1 APGK01023097 KB740463 ENN80600.1 JTDY01007073 KOB65475.1 BABH01029921 KQ460045 KPJ18182.1 KQ459604 KPI92309.1 PYGN01002835 PSN29911.1 KQ434809 KZC06254.1 ODYU01009039 SOQ53153.1 NWSH01000091 PCG79688.1 KQ414667 KOC64664.1 AGBW02013634 OWR43061.1 GL450687 EFN80645.1 KZ288194 PBC33869.1 KQ435848 KOX71079.1 JN159476 AET21260.1 GEZM01046475 GEZM01046474 GEZM01046473 JAV77211.1 KQ971351 EFA06542.1 APGK01057807 KB741282 KB631802 ENN70804.1 ERL86256.1 GBHO01030567 JAG13037.1 KQ980734 KYN13801.1 KQ981305 KYN42937.1 ADTU01009973 GL888529 EGI59738.1 AAZO01003886 DS235339 EEB14914.1 PYGN01000730 PSN41587.1 KK107119 EZA58692.1 GECU01015517 JAS92189.1 GBYB01004932 JAG74699.1 KQ976455 KYM85117.1 GECZ01019434 JAS50335.1 GFTR01005875 JAW10551.1 GL766762 EFZ14632.1 GEBQ01002223 JAT37754.1 KQ982686 KYQ52330.1 UFQS01001902 UFQT01001902 SSX12670.1 SSX32113.1 CH964272 EDW84522.1 CH940652 EDW59340.1 CH902617 EDV42663.1 KOB76863.1 CP012526 ALC46309.1 ALC46310.1 CM000070 EAL27672.1 CH479179 EDW24830.1 OUUW01000013 SPP87984.1 CH954181 EDV48586.1 CH916369 EDV93282.1 CH933806 EDW14423.1 CM000160 EDW95920.1 KRK02799.1 JXJN01028586 CCAG010005923 AE014297 AY089432 AAF55571.1 AAL90170.1 CM000364 EDX12435.1 GAMC01012362 JAB94193.1 CH480820 EDW54632.1
KPJ01853.1 NWSH01003604 PCG66067.1 KQ460761 KPJ12523.1 ODYU01000577 SOQ35597.1 AAZO01005497 DS235786 EEB17054.1 KK107372 QOIP01000004 EZA51705.1 RLU23740.1 GL762841 EFZ20670.1 KQ764887 OAD54258.1 GL888932 EGI57207.1 KZ288439 PBC25730.1 JTDY01004102 KOB68598.1 KQ977059 KYN06039.1 GL434492 EFN74843.1 GBYB01015269 JAG85036.1 KQ980581 KYN15282.1 KQ982373 KYQ56993.1 KQ435826 KOX71989.1 ADTU01011290 LBMM01004298 KMQ92593.1 KQ976500 KYM83202.1 KQ971338 KYB28052.1 OWR55453.1 SOQ35598.1 KPJ12526.1 PCG66066.1 KPJ01854.1 KQ414670 KOC64434.1 KQ434857 KZC08871.1 LJIJ01000133 ODN01930.1 UFQT01000658 SSX26220.1 UFQS01000871 UFQT01000871 SSX07413.1 SSX27755.1 APGK01023097 KB740463 ENN80600.1 JTDY01007073 KOB65475.1 BABH01029921 KQ460045 KPJ18182.1 KQ459604 KPI92309.1 PYGN01002835 PSN29911.1 KQ434809 KZC06254.1 ODYU01009039 SOQ53153.1 NWSH01000091 PCG79688.1 KQ414667 KOC64664.1 AGBW02013634 OWR43061.1 GL450687 EFN80645.1 KZ288194 PBC33869.1 KQ435848 KOX71079.1 JN159476 AET21260.1 GEZM01046475 GEZM01046474 GEZM01046473 JAV77211.1 KQ971351 EFA06542.1 APGK01057807 KB741282 KB631802 ENN70804.1 ERL86256.1 GBHO01030567 JAG13037.1 KQ980734 KYN13801.1 KQ981305 KYN42937.1 ADTU01009973 GL888529 EGI59738.1 AAZO01003886 DS235339 EEB14914.1 PYGN01000730 PSN41587.1 KK107119 EZA58692.1 GECU01015517 JAS92189.1 GBYB01004932 JAG74699.1 KQ976455 KYM85117.1 GECZ01019434 JAS50335.1 GFTR01005875 JAW10551.1 GL766762 EFZ14632.1 GEBQ01002223 JAT37754.1 KQ982686 KYQ52330.1 UFQS01001902 UFQT01001902 SSX12670.1 SSX32113.1 CH964272 EDW84522.1 CH940652 EDW59340.1 CH902617 EDV42663.1 KOB76863.1 CP012526 ALC46309.1 ALC46310.1 CM000070 EAL27672.1 CH479179 EDW24830.1 OUUW01000013 SPP87984.1 CH954181 EDV48586.1 CH916369 EDV93282.1 CH933806 EDW14423.1 CM000160 EDW95920.1 KRK02799.1 JXJN01028586 CCAG010005923 AE014297 AY089432 AAF55571.1 AAL90170.1 CM000364 EDX12435.1 GAMC01012362 JAB94193.1 CH480820 EDW54632.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000053268
UP000218220
UP000053240
+ More
UP000009046 UP000053097 UP000279307 UP000192223 UP000007755 UP000005203 UP000242457 UP000078542 UP000000311 UP000078492 UP000075809 UP000053105 UP000005205 UP000036403 UP000078540 UP000007266 UP000053825 UP000076502 UP000094527 UP000019118 UP000245037 UP000008237 UP000030742 UP000078541 UP000079169 UP000095300 UP000007798 UP000008792 UP000007801 UP000095301 UP000192221 UP000092553 UP000001819 UP000008744 UP000268350 UP000008711 UP000001070 UP000009192 UP000091820 UP000002282 UP000092460 UP000092444 UP000092443 UP000092445 UP000000803 UP000000304 UP000001292
UP000009046 UP000053097 UP000279307 UP000192223 UP000007755 UP000005203 UP000242457 UP000078542 UP000000311 UP000078492 UP000075809 UP000053105 UP000005205 UP000036403 UP000078540 UP000007266 UP000053825 UP000076502 UP000094527 UP000019118 UP000245037 UP000008237 UP000030742 UP000078541 UP000079169 UP000095300 UP000007798 UP000008792 UP000007801 UP000095301 UP000192221 UP000092553 UP000001819 UP000008744 UP000268350 UP000008711 UP000001070 UP000009192 UP000091820 UP000002282 UP000092460 UP000092444 UP000092443 UP000092445 UP000000803 UP000000304 UP000001292
Interpro
IPR000719
Prot_kinase_dom
+ More
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR016181 Acyl_CoA_acyltransferase
IPR013523 Hist_AcTrfase_HAT1_C
IPR037113 Hat1_N_sf
IPR019467 Hat1_N
IPR033898 RNAP_AC19
IPR036603 RBP11-like
IPR008193 RNA_pol_Rpb11_13-16kDa_CS
IPR009025 RBP11-like_dimer
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR016181 Acyl_CoA_acyltransferase
IPR013523 Hist_AcTrfase_HAT1_C
IPR037113 Hat1_N_sf
IPR019467 Hat1_N
IPR033898 RNAP_AC19
IPR036603 RBP11-like
IPR008193 RNA_pol_Rpb11_13-16kDa_CS
IPR009025 RBP11-like_dimer
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
Gene 3D
ProteinModelPortal
H9IUT3
A0A0L7LN37
A0A212FNY0
A0A194Q8K1
A0A2A4J1M8
A0A194R3Y9
+ More
A0A2H1V438 E0VUJ8 A0A026W6M0 E9IFJ5 A0A310SBN0 A0A1W4XJG4 F4X876 A0A087ZQW3 A0A2A3E208 A0A0L7KZB1 A0A151IMD4 E1ZVD3 A0A0C9RFD0 A0A195DR43 A0A151X9F2 A0A0M8ZY63 A0A158NC21 A0A0J7KQI0 A0A195BG99 A0A139WJG5 H9IUT2 A0A212FP11 A0A2H1V427 A0A194R9I7 A0A2A4J2V2 A0A194Q8W4 A0A0L7R0R0 A0A154PAK5 A0A1D2N9J0 A0A336M858 A0A336MBU8 N6UFU9 A0A0L7KQT7 H9J602 A0A194RKW2 A0A194PHV9 A0A2P8XD25 A0A154P4L0 A0A2H1WJA0 A0A2A4K6Z1 A0A0L7R1E4 A0A212ENN5 E2BUI7 A0A2A3ERW8 A0A088AFP6 A0A0M8ZWN2 G8GJE9 A0A1Y1LVT0 D6WR20 N6SSY9 A0A0A9WX60 A0A195DLT1 A0A195FRM3 A0A158NAR6 F4X1E3 E0VNF8 A0A2P8YBE4 A0A026WRP4 A0A1B6IZ62 A0A3Q0IUA1 A0A0C9QW83 A0A1S3D243 A0A195BKA0 A0A1B6FJH7 A0A224XRB7 E9IXN5 A0A1B6MP88 A0A151WWS3 A0A1I8PN93 A0A336MTK5 B4NH25 B4M3X9 B3LW85 A0A0L7LMS7 A0A1I8MVU7 A0A1W4UJJ2 A0A0M3QXR1 A0A0M4EQ10 Q299N1 B4G5G4 A0A3B0K176 B3NZ44 B4JF01 B4K8M4 A0A1A9W2S2 B4PLY8 A0A1B0C7H9 A0A1B0FEQ0 A0A1A9Y9F6 A0A1A9ZE95 Q9VE58 B4QU12 W8BB14 B4I275
A0A2H1V438 E0VUJ8 A0A026W6M0 E9IFJ5 A0A310SBN0 A0A1W4XJG4 F4X876 A0A087ZQW3 A0A2A3E208 A0A0L7KZB1 A0A151IMD4 E1ZVD3 A0A0C9RFD0 A0A195DR43 A0A151X9F2 A0A0M8ZY63 A0A158NC21 A0A0J7KQI0 A0A195BG99 A0A139WJG5 H9IUT2 A0A212FP11 A0A2H1V427 A0A194R9I7 A0A2A4J2V2 A0A194Q8W4 A0A0L7R0R0 A0A154PAK5 A0A1D2N9J0 A0A336M858 A0A336MBU8 N6UFU9 A0A0L7KQT7 H9J602 A0A194RKW2 A0A194PHV9 A0A2P8XD25 A0A154P4L0 A0A2H1WJA0 A0A2A4K6Z1 A0A0L7R1E4 A0A212ENN5 E2BUI7 A0A2A3ERW8 A0A088AFP6 A0A0M8ZWN2 G8GJE9 A0A1Y1LVT0 D6WR20 N6SSY9 A0A0A9WX60 A0A195DLT1 A0A195FRM3 A0A158NAR6 F4X1E3 E0VNF8 A0A2P8YBE4 A0A026WRP4 A0A1B6IZ62 A0A3Q0IUA1 A0A0C9QW83 A0A1S3D243 A0A195BKA0 A0A1B6FJH7 A0A224XRB7 E9IXN5 A0A1B6MP88 A0A151WWS3 A0A1I8PN93 A0A336MTK5 B4NH25 B4M3X9 B3LW85 A0A0L7LMS7 A0A1I8MVU7 A0A1W4UJJ2 A0A0M3QXR1 A0A0M4EQ10 Q299N1 B4G5G4 A0A3B0K176 B3NZ44 B4JF01 B4K8M4 A0A1A9W2S2 B4PLY8 A0A1B0C7H9 A0A1B0FEQ0 A0A1A9Y9F6 A0A1A9ZE95 Q9VE58 B4QU12 W8BB14 B4I275
PDB
2R0I
E-value=1.12794e-37,
Score=390
Ontologies
GO
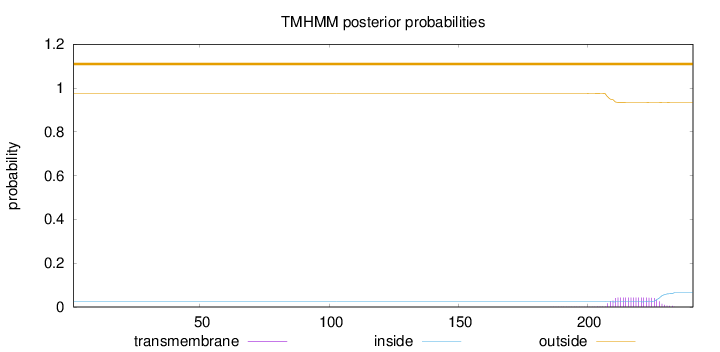
Topology
Length:
241
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.8714
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02424
outside
1 - 241
Population Genetic Test Statistics
Pi
161.351739
Theta
126.047203
Tajima's D
1.009662
CLR
0.494463
CSRT
0.667016649167542
Interpretation
Uncertain
