Pre Gene Modal
BGIBMGA001011
Annotation
PREDICTED:_ER_degradation-enhancing_alpha-mannosidase-like_protein_2_isoform_X2_[Bombyx_mori]
Full name
alpha-1,2-Mannosidase
Location in the cell
Cytoplasmic Reliability : 1.363 Nuclear Reliability : 1.379 PlasmaMembrane Reliability : 1.301
Sequence
CDS
ATGGTCAAATTAACCTTCCTGATCAGTATTTCAGTGTTGACTTGTAATTTAATTTTAAGTGTACGAGTATACAAACGTGCAGATATACTTAAACTAAGAGAGGAAGTACGTGAAATGTTTCAACATGCATACGACAGCTATTTAAGATATGCATATCCTTATGACGAATTACGTCCATTGAGCTGCGACGGTGTTGATACCTGGGGCAGTTACTCACTCACCCTCATTGATGCATTGGACACGCTTGCCATTATGGGAAACTATAGTGAATTTAATCGAGTAGTAGATATAGTACTACAGAAAAGACACTTTGATGCAGACATTAACGTTTCAGTGTTTGAGACGAATATTAGAATTGTTGGCGGTTTATTGAGTGCTCATTTACTGTCACATAAAACTGGGATGAAATTAGAACCAGGCTGGCCATGCAATGGTCCTTTACTTCGTTTAGCTGAAGATGTAGCACAGAGACTAATAGCTGCATTTGATACAACTACTGGAATGCCTTATGGAACAATTAACTTGCGATCAGGTGTTCCACCTGGGGAAACTAGTATCACATGCACTGCTGGAGTTGGTACATTTATAATTGAGTTTGGTACTTTGAGTAGACTGACAGGAGATCCATTGTATGAAGAGGTAGCTTATAATGCTTTGAAAGCCTTATACCATCATAGATCACCAATTGGTCTCCTTGGCAACCACATAGATGTGATGACTGGACGCTGGACAGCTCAAGATGCTGGAATTGGTGGTGGAATCGATTCTTATTATGAATATTTGGTTAAAGGTGCCATACTACTAGAGAAGCCAGAGTTAATGTCCATGTTCTTAGAAGCACGTCAATCTATTGAGAAGTACCTGAAAAAAGATGATTGGTTTGTGTGGGCCACTATGTTGCGTGGGCACGTCACCTTACCTGTCTTTCAGTCTCTTGAATCATATTGGCCAGGTCTTTTAAGCCTCATAGGCGAAAGTGACGCAGCGATGCGCATAATTCACAACTACCACAGCGTGTGGCGCCAATATGGATTTACACCTGAAGTGTACAATCTCGGGACGGGTGAGGCGTCTTCGTCACGCGAGAGTTATCCCCTACGACCCGAACTCATAGAGTCAATCATGTATTTGTACCGAGACACCAGAGACCCTATACTGTTGCAAATGGGCGAAGATATTCTGAGGAGCATACAGCATAGTGCCAGGACACCGTGTGGGTATGCAACGATAAAAGATGTTCGTGACCATCGTAAAGAAGATCGCATGGAATCGTTTTTCTTAGCCGAAACCACAAAATACCTGTACCTACTGTTCGATCCGGATAATTTCATTCACAACCCTGGGGTCCGCGGTACTGTCATCGACACACCTAATGGCGAATGCGTGGTCGACCTTGGCGGATACATCTTCAATACAGAAGCACATCCGATTGACCCTAACATGCTGTATTGCTGCCATGAAGCTAGACAAGGTATAAATATCAGCGAGGTTTACAGTATATACCAAATTTTGGAAGAAGAAGATAATATAAAATTTATGACTATGATAGAAGCGAACACCAATGAAACTAAGATCACTCAATCGAACAACAACAATAATGAGAGTGCAATAGAAAATAAAACAATTGACCACACAGAACCGAAGGAAAGATCAGATGAAAACGAGCCTGAACAAAATCAAGTATTGATAAAAACTGAATCGAGAACCGGATACGTCAACCTCGACGGAACGGAAAGGGAGAACGCAAACGAATCAAACCTCAGTGATCTTGTTAAAGAGACGATCAATAAATATTACAACGACACTATAAATGCCGAAACACAAGAAAATGTTAAACATTCAGCCGAAGAACCAACGGCAGGATCGTCGCAAACCGTTGAAGTGGACGACATAATCGTGCCAGAGAAATCGGAGGTGCAACTGCCAAGCGTCACGAAAACGAAAGAAGTAATGAACATGCTGCCAAAAGTTATACAAGACTTTTTGAACAGCGATTGGAAATCGAAACCGAAATGCGAACCGCAGAAGATGTTAGAAAGGATACGTAGAGAACAGAAGTATCCAGATCACCCGGATGTATATAAGTACGAGTTATTATTGACGCCGGCTCCGTCGTTTCTACAAAGAATATCGTTGGCCGGAGAGTTCTTAAATAAGAAGCAACTGGATAAAGAAGATTCACATAAATCAACGCTTGCTTGTAAAATTATTGAAACGGCAAAGGCCCCAAAAGATTTTGTTGTAAAATTTTTGCCACGCGAAATAGATGTGCTCGTGCGCTTGAATCATCCGCATCTGATTCACGTGCATAGCATATTTCAGAGGAAAACAAAATACTATATTTTCATGAGGTACACGGAAAATGGTGATTTACTGAGTTATATACTAAAAAACGGGTGCGTGTCTGAAAATCAAGCTCGCGTCTGGACTCGACAACTTGCATTGGGACTACAATACTTGCACGAGTTAGAAATCGCTCACAGAGACATCAAATGCGAGAACGTACTGTTGACGGCGAATTTTAACGTGAAATTATCTGATTTTGGCTTTTCCCGTTTCTGTGTTGAAAGCGACAATCAGCCTGTCTTAAGCGAAACTTATTGCGGTTCTATGTCATACGCTGCACCAGAAATACTACGCGGAAAACCGTATTGCCCGAAGCCAACGGATTTATGGTCCTTAGGCGTCGTACTATTTGTAATGCTGAACAAATCAATGCCATTTGACGACACGCGTATGAGAAAACTTTATGAACAGCAAATGGGAAAGAAATATCGCTTTAGATCGCGCGTAGCTAGTATACTCTCGCTCGAATGTAAGACTGTTGTAAAGCATTTGTTAGAACCGGATCCAGGACTCCGACATTCCGCTACCAATGTTCTCGACTCTGAATGGATTGCTATGGATAGTAGATTAACCAGTAAGTATAGAAATATATTAAATACTAGCTGA
Protein
MVKLTFLISISVLTCNLILSVRVYKRADILKLREEVREMFQHAYDSYLRYAYPYDELRPLSCDGVDTWGSYSLTLIDALDTLAIMGNYSEFNRVVDIVLQKRHFDADINVSVFETNIRIVGGLLSAHLLSHKTGMKLEPGWPCNGPLLRLAEDVAQRLIAAFDTTTGMPYGTINLRSGVPPGETSITCTAGVGTFIIEFGTLSRLTGDPLYEEVAYNALKALYHHRSPIGLLGNHIDVMTGRWTAQDAGIGGGIDSYYEYLVKGAILLEKPELMSMFLEARQSIEKYLKKDDWFVWATMLRGHVTLPVFQSLESYWPGLLSLIGESDAAMRIIHNYHSVWRQYGFTPEVYNLGTGEASSSRESYPLRPELIESIMYLYRDTRDPILLQMGEDILRSIQHSARTPCGYATIKDVRDHRKEDRMESFFLAETTKYLYLLFDPDNFIHNPGVRGTVIDTPNGECVVDLGGYIFNTEAHPIDPNMLYCCHEARQGINISEVYSIYQILEEEDNIKFMTMIEANTNETKITQSNNNNNESAIENKTIDHTEPKERSDENEPEQNQVLIKTESRTGYVNLDGTERENANESNLSDLVKETINKYYNDTINAETQENVKHSAEEPTAGSSQTVEVDDIIVPEKSEVQLPSVTKTKEVMNMLPKVIQDFLNSDWKSKPKCEPQKMLERIRREQKYPDHPDVYKYELLLTPAPSFLQRISLAGEFLNKKQLDKEDSHKSTLACKIIETAKAPKDFVVKFLPREIDVLVRLNHPHLIHVHSIFQRKTKYYIFMRYTENGDLLSYILKNGCVSENQARVWTRQLALGLQYLHELEIAHRDIKCENVLLTANFNVKLSDFGFSRFCVESDNQPVLSETYCGSMSYAAPEILRGKPYCPKPTDLWSLGVVLFVMLNKSMPFDDTRMRKLYEQQMGKKYRFRSRVASILSLECKTVVKHLLEPDPGLRHSATNVLDSEWIAMDSRLTSKYRNILNTS
Summary
Similarity
Belongs to the glycosyl hydrolase 47 family.
Feature
chain alpha-1,2-Mannosidase
Uniprot
EC Number
3.2.1.-
Pubmed
EMBL
ODYU01000577
SOQ35599.1
KQ460761
KPJ12527.1
KQ459299
KPJ01855.1
+ More
AGBW02004200 OWR55452.1 GEZM01099009 JAV53576.1 AJWK01021254 ACPB03022213 KZ288186 PBC34746.1 KQ981799 KYN35691.1 KQ979074 KYN22858.1 KQ982651 KYQ52838.1 KK107921 QOIP01000001 EZA47245.1 RLU26311.1 KK852793 KDR16453.1 GL888262 EGI63661.1 KQ977586 KYN01682.1 GL766449 EFZ15264.1 GL441177 EFN65077.1 KQ435876 KOX70169.1 KQ414666 KOC65029.1
AGBW02004200 OWR55452.1 GEZM01099009 JAV53576.1 AJWK01021254 ACPB03022213 KZ288186 PBC34746.1 KQ981799 KYN35691.1 KQ979074 KYN22858.1 KQ982651 KYQ52838.1 KK107921 QOIP01000001 EZA47245.1 RLU26311.1 KK852793 KDR16453.1 GL888262 EGI63661.1 KQ977586 KYN01682.1 GL766449 EFZ15264.1 GL441177 EFN65077.1 KQ435876 KOX70169.1 KQ414666 KOC65029.1
Proteomes
Pfam
PF01532 Glyco_hydro_47
Interpro
SUPFAM
SSF48225
SSF48225
Gene 3D
ProteinModelPortal
PDB
5KKB
E-value=8.4166e-57,
Score=562
Ontologies
GO
Topology
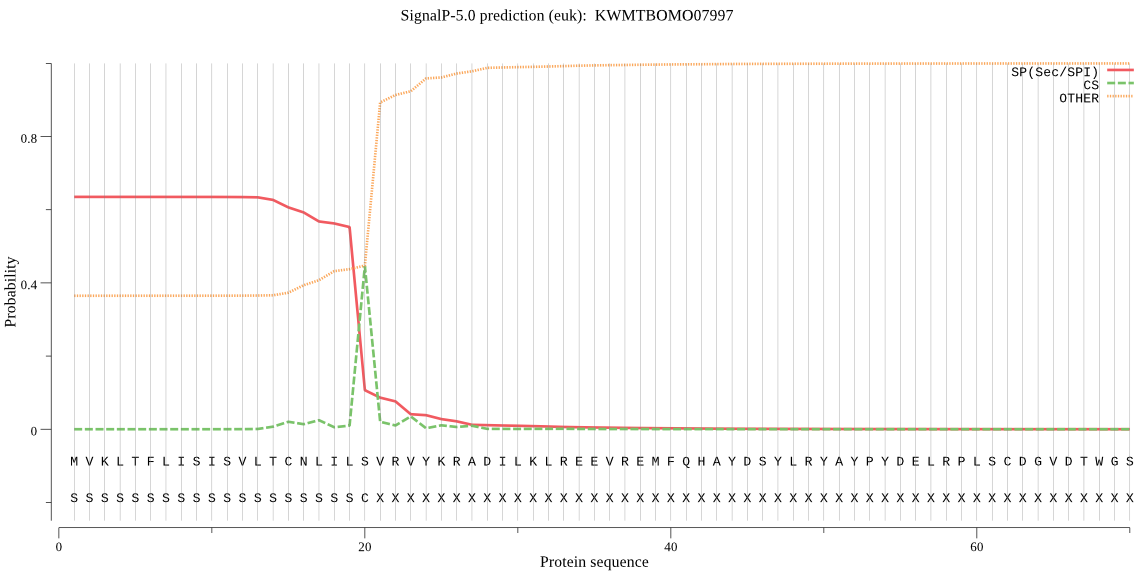
SignalP
Position: 1 - 20,
Likelihood: 0.634926
Length:
983
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.36507999999997
Exp number, first 60 AAs:
6.54729
Total prob of N-in:
0.30394
outside
1 - 983

Population Genetic Test Statistics
Pi
160.696083
Theta
170.543261
Tajima's D
0
CLR
32.645811
CSRT
0.367381630918454
Interpretation
Uncertain
