Gene
KWMTBOMO07996
Pre Gene Modal
BGIBMGA001142
Annotation
cyclin_A_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.674
Sequence
CDS
ATGGCTTTTAGAATTCATGAAGATCAAGAAAACACAGCATTAGGCCTAAGAAAAGATGGATCCGACCTATTTTCTACCAACCAAAGGCGTGCCCTCGGAGACCTCAGTCAATTTGCATGCAACCATAATCGGAACACCAAACTAACTGGTCTGACAAACGGAACTTGTAAGGTTCAAGATGAGAACAGGACGGTACGTCAAATCAAGAACGAGAAAAACATCGTTCTTCCCCTATCCCAGTTTCGTGCCTTTAGCGTATATGAAGACAAACCCACCGAGGTGGAGGTGAAAAAGCGGGAGACCACTTACAAACCATTTGCAACCAAGGAGACTAAAAAAGAAAGCAATTTCTTTGTGAATGCTGCTGAAAATGTGAGAGCTCTGTGTGCACAAGTTGAAAAAAAATCGCTTGAGGAAACCAAACCAGATCGATCGAGAGAAGAACCCCCATTACGGAAAGCATTACAAGAAAAGAAAGATGCTGTTGAATCACCCATGTCTGTGGTGGATGCCAGTATTCTGTCGATGTCTATTTCAAAGAATGAGAGTCAAATTATTGAAGATGTTGACGATGAAGAAATCACCACTGCACAGACCGACAGAGAAATGTTTTTCTATGTTGAGGAATATAGGCAGGATATCTATGAATACATGAGGGAAATTGAGGTGAAAAACAGGGCCAACCCCCGCTACATGCGTAAGCAACCAGACATCACACATGTTATGCGCTCAATACTTGTGGACTGGCTAGTTGAAGTATGTGATGAATATCAACAGCAGAGTGAAACTTTACATCTAGCTGTGTCTTACGTAGATAGGTTCCTGTCATACATGAGTGTTGTCAGAACTAAGCTTCAACTTGTCGGAACAGCTGCCACCTATATTGCCGCTAAATATGAAGAAGTATATCCACCAGAAGTCTCAGAATTTGTGTACATCACTGACGACACATACACAAAACGTGAAGTACTGAGAATGGAGCATTTAATTTTAAAGGTGTTATCATTTGATCTCTCAACACCCACCTCACTAGCTTTCCTGTCACATTACTGTATTTCTAATGGTCTCTCAAAGAAGACATTCCATTTGGCATCTTATATTGCGGAGCTGTGCCTACTTGAAGCAGACCCATATCTTCAATTCAAACCCTCAGTAATAGCAGCGTCGGCGCTGGCAACGGCTCGGCATTGCTTACTATGCGAGCAGTGCGCCTGTGATCCTCAGGACGTATACGAAACTAGAGACGCTCCGGGCAAGGTCAACCCGCAATGTGCAATGGTCGCGTGGCCGTCTACGTTGTCCACGTGTTCGGGATACACTTTACTTGAATTGGAAACATGCCTTAAAGAAATTGCTCGTACACATTCCCATGCCTCTGTTCAACCTTATCAAGCCATACCAGACAAATATAAGAGCAACAAGTTTGAAGGCGTGTCACAAGTAGAGCCGCGGCCCATGTTCCCGGTGGGCAAGTACCAAGCCCCGGCCGCCGCCCGCCCGCCCCCAGCGGCCGACTCTGCGCGCGCCACCAGCTAG
Protein
MAFRIHEDQENTALGLRKDGSDLFSTNQRRALGDLSQFACNHNRNTKLTGLTNGTCKVQDENRTVRQIKNEKNIVLPLSQFRAFSVYEDKPTEVEVKKRETTYKPFATKETKKESNFFVNAAENVRALCAQVEKKSLEETKPDRSREEPPLRKALQEKKDAVESPMSVVDASILSMSISKNESQIIEDVDDEEITTAQTDREMFFYVEEYRQDIYEYMREIEVKNRANPRYMRKQPDITHVMRSILVDWLVEVCDEYQQQSETLHLAVSYVDRFLSYMSVVRTKLQLVGTAATYIAAKYEEVYPPEVSEFVYITDDTYTKREVLRMEHLILKVLSFDLSTPTSLAFLSHYCISNGLSKKTFHLASYIAELCLLEADPYLQFKPSVIAASALATARHCLLCEQCACDPQDVYETRDAPGKVNPQCAMVAWPSTLSTCSGYTLLELETCLKEIARTHSHASVQPYQAIPDKYKSNKFEGVSQVEPRPMFPVGKYQAPAAARPPPAADSARATS
Summary
Similarity
Belongs to the cyclin family.
Uniprot
C0KLC6
D3KYT0
H9IV62
A0A2A4J5X0
A0A2H1V430
A0A159ZM63
+ More
A0A194QEG5 A0A194R3Z2 S4NXS6 A0A212FNY1 A0A0L7KZE5 A0A232FEB5 K7IYS2 A0A1W4X1G6 D2A0W3 A0A067RLB0 A0A0C9QU55 A0A291S6U8 A0A2A3EQK3 U4UL13 A0A087ZRM6 A0A0N0U635 A0A1Q3FVN6 A0A2J7PNN2 A0A1Q3FVI2 A0A1Y1MAX6 A0A195FLS6 A0A195BUT2 A0A151XJ02 A0A182JQ41 F4WRV2 A0A154PFY7 Q17NN3 A0A023ET01 A0A182WZU3 A0A182GFJ6 A0A182MYT8 A0A195ENW5 A0A310SWU3 E9ID93 A0A0J7LAC7 E2AFN5 A0A182HQD8 A0A195BZU4 A0A182KSA3 A0A182VFP5 A0A182WEW9 A0A1B6LX76 E2B3E7 A0A1I8PRV0 A0A1I8N208 W5JK10 A0A182P5U9 A0A182RD17 E9HA37 A0A1B0GDC4 A0A0N7ZYA6 A0A0P5FKC9 A0A0P5AD35 A0A0N8A4L9 A0A0P5EHE5 A0A0P5T704 A0A0P5VFS4
A0A194QEG5 A0A194R3Z2 S4NXS6 A0A212FNY1 A0A0L7KZE5 A0A232FEB5 K7IYS2 A0A1W4X1G6 D2A0W3 A0A067RLB0 A0A0C9QU55 A0A291S6U8 A0A2A3EQK3 U4UL13 A0A087ZRM6 A0A0N0U635 A0A1Q3FVN6 A0A2J7PNN2 A0A1Q3FVI2 A0A1Y1MAX6 A0A195FLS6 A0A195BUT2 A0A151XJ02 A0A182JQ41 F4WRV2 A0A154PFY7 Q17NN3 A0A023ET01 A0A182WZU3 A0A182GFJ6 A0A182MYT8 A0A195ENW5 A0A310SWU3 E9ID93 A0A0J7LAC7 E2AFN5 A0A182HQD8 A0A195BZU4 A0A182KSA3 A0A182VFP5 A0A182WEW9 A0A1B6LX76 E2B3E7 A0A1I8PRV0 A0A1I8N208 W5JK10 A0A182P5U9 A0A182RD17 E9HA37 A0A1B0GDC4 A0A0N7ZYA6 A0A0P5FKC9 A0A0P5AD35 A0A0N8A4L9 A0A0P5EHE5 A0A0P5T704 A0A0P5VFS4
Pubmed
EMBL
FJ619105
ACM79367.1
AB546966
BAI77429.1
BABH01000558
NWSH01003112
+ More
PCG66924.1 ODYU01000577 SOQ35600.1 KU051540 AMY96431.1 KQ459299 KPJ01856.1 KQ460761 KPJ12528.1 GAIX01012042 JAA80518.1 AGBW02004200 OWR55451.1 JTDY01004102 KOB68597.1 NNAY01000335 OXU29124.1 KQ971338 EFA01624.1 KK852444 KDR23813.1 GBYB01004202 JAG73969.1 MF432986 ATL75337.1 KZ288202 PBC33506.1 KB632374 ERL93807.1 KQ435740 KOX76832.1 GFDL01003437 JAV31608.1 NEVH01023380 PNF17945.1 GFDL01003436 JAV31609.1 GEZM01040004 JAV81016.1 KQ981490 KYN41202.1 KQ976401 KYM92379.1 KQ982080 KYQ60317.1 GL888292 EGI63109.1 KQ434886 KZC10100.1 CH477198 EAT48280.1 GAPW01001175 JAC12423.1 JXUM01059639 JXUM01059640 KQ562064 KXJ76764.1 KQ978625 KYN29589.1 KQ759874 OAD62364.1 GL762454 EFZ21530.1 LBMM01000137 KMR04793.1 GL439118 EFN67786.1 APCN01003311 KQ978501 KYM93446.1 GEBQ01011679 JAT28298.1 GL445323 EFN89825.1 ADMH02001279 ETN63235.1 GL732610 EFX71416.1 CCAG010004719 CCAG010004720 GDIP01200689 JAJ22713.1 GDIP01147150 JAJ76252.1 GDIP01200688 JAJ22714.1 GDIP01182985 JAJ40417.1 GDIP01209943 GDIP01148453 GDIP01109403 GDIP01049946 LRGB01001581 JAJ74949.1 KZS11122.1 GDIP01130096 JAL73618.1 GDIP01100537 JAM03178.1
PCG66924.1 ODYU01000577 SOQ35600.1 KU051540 AMY96431.1 KQ459299 KPJ01856.1 KQ460761 KPJ12528.1 GAIX01012042 JAA80518.1 AGBW02004200 OWR55451.1 JTDY01004102 KOB68597.1 NNAY01000335 OXU29124.1 KQ971338 EFA01624.1 KK852444 KDR23813.1 GBYB01004202 JAG73969.1 MF432986 ATL75337.1 KZ288202 PBC33506.1 KB632374 ERL93807.1 KQ435740 KOX76832.1 GFDL01003437 JAV31608.1 NEVH01023380 PNF17945.1 GFDL01003436 JAV31609.1 GEZM01040004 JAV81016.1 KQ981490 KYN41202.1 KQ976401 KYM92379.1 KQ982080 KYQ60317.1 GL888292 EGI63109.1 KQ434886 KZC10100.1 CH477198 EAT48280.1 GAPW01001175 JAC12423.1 JXUM01059639 JXUM01059640 KQ562064 KXJ76764.1 KQ978625 KYN29589.1 KQ759874 OAD62364.1 GL762454 EFZ21530.1 LBMM01000137 KMR04793.1 GL439118 EFN67786.1 APCN01003311 KQ978501 KYM93446.1 GEBQ01011679 JAT28298.1 GL445323 EFN89825.1 ADMH02001279 ETN63235.1 GL732610 EFX71416.1 CCAG010004719 CCAG010004720 GDIP01200689 JAJ22713.1 GDIP01147150 JAJ76252.1 GDIP01200688 JAJ22714.1 GDIP01182985 JAJ40417.1 GDIP01209943 GDIP01148453 GDIP01109403 GDIP01049946 LRGB01001581 JAJ74949.1 KZS11122.1 GDIP01130096 JAL73618.1 GDIP01100537 JAM03178.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000215335 UP000002358 UP000192223 UP000007266 UP000027135 UP000242457 UP000030742 UP000005203 UP000053105 UP000235965 UP000078541 UP000078540 UP000075809 UP000075881 UP000007755 UP000076502 UP000008820 UP000076407 UP000069940 UP000249989 UP000075884 UP000078492 UP000036403 UP000000311 UP000075840 UP000078542 UP000075882 UP000075903 UP000075920 UP000008237 UP000095300 UP000095301 UP000000673 UP000075885 UP000075900 UP000000305 UP000092444 UP000076858
UP000215335 UP000002358 UP000192223 UP000007266 UP000027135 UP000242457 UP000030742 UP000005203 UP000053105 UP000235965 UP000078541 UP000078540 UP000075809 UP000075881 UP000007755 UP000076502 UP000008820 UP000076407 UP000069940 UP000249989 UP000075884 UP000078492 UP000036403 UP000000311 UP000075840 UP000078542 UP000075882 UP000075903 UP000075920 UP000008237 UP000095300 UP000095301 UP000000673 UP000075885 UP000075900 UP000000305 UP000092444 UP000076858
Interpro
SUPFAM
SSF47954
SSF47954
CDD
ProteinModelPortal
C0KLC6
D3KYT0
H9IV62
A0A2A4J5X0
A0A2H1V430
A0A159ZM63
+ More
A0A194QEG5 A0A194R3Z2 S4NXS6 A0A212FNY1 A0A0L7KZE5 A0A232FEB5 K7IYS2 A0A1W4X1G6 D2A0W3 A0A067RLB0 A0A0C9QU55 A0A291S6U8 A0A2A3EQK3 U4UL13 A0A087ZRM6 A0A0N0U635 A0A1Q3FVN6 A0A2J7PNN2 A0A1Q3FVI2 A0A1Y1MAX6 A0A195FLS6 A0A195BUT2 A0A151XJ02 A0A182JQ41 F4WRV2 A0A154PFY7 Q17NN3 A0A023ET01 A0A182WZU3 A0A182GFJ6 A0A182MYT8 A0A195ENW5 A0A310SWU3 E9ID93 A0A0J7LAC7 E2AFN5 A0A182HQD8 A0A195BZU4 A0A182KSA3 A0A182VFP5 A0A182WEW9 A0A1B6LX76 E2B3E7 A0A1I8PRV0 A0A1I8N208 W5JK10 A0A182P5U9 A0A182RD17 E9HA37 A0A1B0GDC4 A0A0N7ZYA6 A0A0P5FKC9 A0A0P5AD35 A0A0N8A4L9 A0A0P5EHE5 A0A0P5T704 A0A0P5VFS4
A0A194QEG5 A0A194R3Z2 S4NXS6 A0A212FNY1 A0A0L7KZE5 A0A232FEB5 K7IYS2 A0A1W4X1G6 D2A0W3 A0A067RLB0 A0A0C9QU55 A0A291S6U8 A0A2A3EQK3 U4UL13 A0A087ZRM6 A0A0N0U635 A0A1Q3FVN6 A0A2J7PNN2 A0A1Q3FVI2 A0A1Y1MAX6 A0A195FLS6 A0A195BUT2 A0A151XJ02 A0A182JQ41 F4WRV2 A0A154PFY7 Q17NN3 A0A023ET01 A0A182WZU3 A0A182GFJ6 A0A182MYT8 A0A195ENW5 A0A310SWU3 E9ID93 A0A0J7LAC7 E2AFN5 A0A182HQD8 A0A195BZU4 A0A182KSA3 A0A182VFP5 A0A182WEW9 A0A1B6LX76 E2B3E7 A0A1I8PRV0 A0A1I8N208 W5JK10 A0A182P5U9 A0A182RD17 E9HA37 A0A1B0GDC4 A0A0N7ZYA6 A0A0P5FKC9 A0A0P5AD35 A0A0N8A4L9 A0A0P5EHE5 A0A0P5T704 A0A0P5VFS4
PDB
4CFU
E-value=3.33254e-69,
Score=666
Ontologies
GO
PANTHER
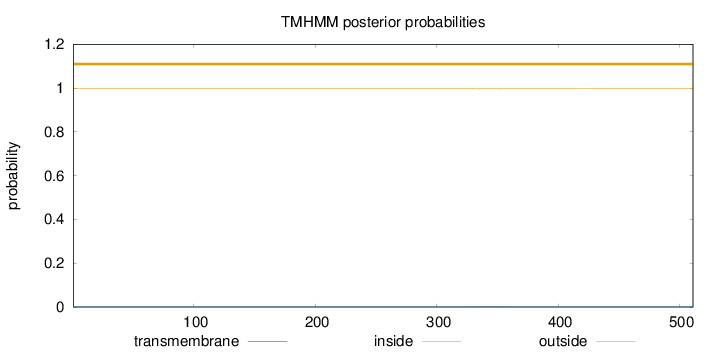
Topology
Length:
511
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03165
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00274
outside
1 - 511
Population Genetic Test Statistics
Pi
137.064208
Theta
127.700272
Tajima's D
-0.582019
CLR
0.343249
CSRT
0.222888855557222
Interpretation
Uncertain
