Gene
KWMTBOMO07992
Annotation
PREDICTED:_uncharacterized_protein_LOC106131122_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.471
Sequence
CDS
ATGGTCAATCAGGAAGTACTTCGAGACTACTCAAGTTCAATCAAAAACTTAAAAAGGGAAGCCGCTAACGCTGGTATTAGTGAAGAAGATTTCAAGAAATTGTATTTTGAATCTCTCAAAACACTAGAAAAAAGTGAACGTCCCAGTTCATTGACAATTGTTAGAAACAATAAACTTAAAATAATTAGCGTACTACTTTTGGTATTTGCTTTATTCAACTTCAAATATGTGTATAGCAGTTTTGTGTGCAACTTACAAGAATACATATATCCAGGACTAAGGTTACTGAGAAAGATCTCAATACCATTTATTTCTTTGTTCCCTGCTTTATCAGAATACTACTATGAAACTTGTTTAGTACAAAACCCATTCTTCACTGTTGTCGATATGGATTGTTGGCCATGTAGTTCTATCACTAATATTCAAGAGATCTTTGATCCAAAACCAGTCACTAATCAGCACATTGCTCCATTTGTTTACCAGACTAGTCAAGAAAGAATTTCAATAAGTCTGCTTAAAAGTCTCTATGAAAAAAATAAAGATATTTTTGCTAATGCATCTCCAAAGATTTTATTAAATAACAAGTTCTATATACAACCAGAAGATATGTTTGATCTACATGCAAAAGGGGAAAAAAATATTTATGTGTGGAAATTCAATCGGATGAATGAAGCAAAAGTGTTACGACAACTTATACCCCGACCGACAGTAGTACCAAAATTTGGACAGAGCATAGAGAGATATTTAATAATACATACTAATCAAGACTCATATAAATTACCAGATACTGAATGTAATTTCTCATTCGTACTATCATTGAGTGGTGCAAGGACACTCTATTTAATACCAGCTGAAGAATGCAAACATCAATGCAGGCATTTGTCAATTGATTTAAAAGAAGGTTATTTACTATGGTACAATTGGTGGTACTGGAGACCAAAAATACAACCTACAGTTGACAATACTACTTTCATTGGCCATATAGGCTCATATTGCTAA
Protein
MVNQEVLRDYSSSIKNLKREAANAGISEEDFKKLYFESLKTLEKSERPSSLTIVRNNKLKIISVLLLVFALFNFKYVYSSFVCNLQEYIYPGLRLLRKISIPFISLFPALSEYYYETCLVQNPFFTVVDMDCWPCSSITNIQEIFDPKPVTNQHIAPFVYQTSQERISISLLKSLYEKNKDIFANASPKILLNNKFYIQPEDMFDLHAKGEKNIYVWKFNRMNEAKVLRQLIPRPTVVPKFGQSIERYLIIHTNQDSYKLPDTECNFSFVLSLSGARTLYLIPAEECKHQCRHLSIDLKEGYLLWYNWWYWRPKIQPTVDNTTFIGHIGSYC
Summary
Uniprot
A0A2A4J4K0
A0A2H1V440
A0A212FNX6
A0A194R9J4
A0A0L7LC29
A0A1Q3F904
+ More
W5JTL5 B0X896 A0A182N9A7 A0A182RJL0 A0A0P6JSP8 A0A1S4FNA9 A0A182WBI3 A0A182JRN7 A0A2M4CNF8 A0A182WWI1 A0A182L3N6 A0A182I6R1 A0A182TZD4 A0A182FEM3 A0A1Y1MH36 A0A139WIX5 A0A182Q8N8 A0A182IQX4 A0A182LTG1 A0A182VAG1 A0A0C9R5D3 A0A0C9RJU9 A0A084W142 A0A1W4XRP4 A0A154PDQ1 A0A026VTY1 A0A1L8DA30 A0A0L7R7A2 E2ACS2 E9J8M0 A0A0K8TKR0 A0A151IES1 Q16V43 A0A182PIJ2 F4W6A4 A0A195FJ32 Q7QIL4 A0A1B6FVE1 A0A232EZW6 A0A2A3E237 U4TUN5 A0A1B6K505 A0A195AW28 E0VCC6 A0A151WHP1 A0A158P220 A0A088A2R4 A0A0M8ZRF4 N6UJH6 A0A195DMM2 A0A1B6MGP5 B4PZ93 B3NXX1 A0A1B6H8I9 B4NEP8 A0A0A9ZFG2 D3TPC9 A0A1B0ABV9 A0A1W4V9R3 A0A1A9UHM6 A0A3B0JWR5 A0A1A9W1W6 A0A1A9XIB8 A0A1B0ARV5 B3MS62 B4JND0 E1JJF2 A0A1B6CT62 A0A1B6CW23 Q29JM5 A0A0K8W4K8 A0A034W9P2 A0A182Y2B6 A0A0A1WLH7 W8AJM0 A0A2J7Q502 T1IB28 A0A1I8NC67 A0A1I8Q8X2 A0A0M3QZA8 B4M7J5 E2BCT9 A0A146LTY3 A0A336ME62 B4L1D2 A0A2M4AY05 A0A2R7VNB8 Q9W3V0 A0A0T6B2F1 A0A2P8XAH9
W5JTL5 B0X896 A0A182N9A7 A0A182RJL0 A0A0P6JSP8 A0A1S4FNA9 A0A182WBI3 A0A182JRN7 A0A2M4CNF8 A0A182WWI1 A0A182L3N6 A0A182I6R1 A0A182TZD4 A0A182FEM3 A0A1Y1MH36 A0A139WIX5 A0A182Q8N8 A0A182IQX4 A0A182LTG1 A0A182VAG1 A0A0C9R5D3 A0A0C9RJU9 A0A084W142 A0A1W4XRP4 A0A154PDQ1 A0A026VTY1 A0A1L8DA30 A0A0L7R7A2 E2ACS2 E9J8M0 A0A0K8TKR0 A0A151IES1 Q16V43 A0A182PIJ2 F4W6A4 A0A195FJ32 Q7QIL4 A0A1B6FVE1 A0A232EZW6 A0A2A3E237 U4TUN5 A0A1B6K505 A0A195AW28 E0VCC6 A0A151WHP1 A0A158P220 A0A088A2R4 A0A0M8ZRF4 N6UJH6 A0A195DMM2 A0A1B6MGP5 B4PZ93 B3NXX1 A0A1B6H8I9 B4NEP8 A0A0A9ZFG2 D3TPC9 A0A1B0ABV9 A0A1W4V9R3 A0A1A9UHM6 A0A3B0JWR5 A0A1A9W1W6 A0A1A9XIB8 A0A1B0ARV5 B3MS62 B4JND0 E1JJF2 A0A1B6CT62 A0A1B6CW23 Q29JM5 A0A0K8W4K8 A0A034W9P2 A0A182Y2B6 A0A0A1WLH7 W8AJM0 A0A2J7Q502 T1IB28 A0A1I8NC67 A0A1I8Q8X2 A0A0M3QZA8 B4M7J5 E2BCT9 A0A146LTY3 A0A336ME62 B4L1D2 A0A2M4AY05 A0A2R7VNB8 Q9W3V0 A0A0T6B2F1 A0A2P8XAH9
Pubmed
22118469
26354079
26227816
20920257
23761445
26999592
+ More
17510324 20966253 28004739 18362917 19820115 24438588 24508170 30249741 20798317 21282665 26369729 21719571 12364791 28648823 23537049 20566863 21347285 17994087 17550304 25401762 26823975 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 25348373 25244985 25830018 24495485 25315136 26109357 26109356 29403074
17510324 20966253 28004739 18362917 19820115 24438588 24508170 30249741 20798317 21282665 26369729 21719571 12364791 28648823 23537049 20566863 21347285 17994087 17550304 25401762 26823975 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 25348373 25244985 25830018 24495485 25315136 26109357 26109356 29403074
EMBL
NWSH01003112
PCG66921.1
ODYU01000577
SOQ35603.1
AGBW02004200
OWR55448.1
+ More
KQ460761 KPJ12531.1 JTDY01001767 KOB72945.1 GFDL01011067 JAV23978.1 ADMH02000413 ETN66623.1 DS232478 EDS42365.1 GDUN01000184 JAN95735.1 GGFL01002678 MBW66856.1 APCN01003693 GEZM01033279 JAV84288.1 KQ971338 KYB27968.1 AXCN02000558 AXCM01000934 GBYB01007987 JAG77754.1 GBYB01013557 GBYB01013558 JAG83324.1 JAG83325.1 ATLV01019208 KE525264 KFB43936.1 KQ434869 KZC09378.1 KK107906 QOIP01000010 EZA47258.1 RLU17654.1 GFDF01010894 JAV03190.1 KQ414642 KOC66719.1 GL438568 EFN68816.1 GL769036 EFZ10812.1 GDAI01002887 JAI14716.1 KQ977858 KYM99313.1 CH477605 EAT38385.1 GL887707 EGI70239.1 KQ981523 KYN40413.1 AAAB01008807 EAA04358.3 GECZ01024567 GECZ01015766 JAS45202.1 JAS54003.1 NNAY01001413 OXU24056.1 KZ288449 PBC25594.1 KB631581 ERL84482.1 GECU01001175 JAT06532.1 KQ976731 KYM76262.1 DS235053 EEB11032.1 KQ983112 KYQ47356.1 ADTU01006966 KQ435951 KOX68113.1 APGK01031920 KB740831 ENN78817.1 KQ980724 KYN14082.1 GEBQ01004926 JAT35051.1 CM000162 EDX01060.1 CH954180 EDV47422.1 GECU01036773 JAS70933.1 CH964239 EDW82217.1 GBHO01022996 GBHO01022995 GBHO01022991 GBHO01022990 GBHO01001436 GBHO01001418 GDHC01015722 JAG20608.1 JAG20609.1 JAG20613.1 JAG20614.1 JAG42168.1 JAG42186.1 JAQ02907.1 CCAG010021577 EZ423281 ADD19557.1 OUUW01000003 SPP77796.1 JXJN01002588 CH902622 EDV34617.1 CH916371 EDV92223.1 AE014298 ACZ95219.1 GEDC01020592 JAS16706.1 GEDC01019592 JAS17706.1 CH379063 EAL32276.2 GDHF01033302 GDHF01006278 JAI19012.1 JAI46036.1 GAKP01007920 JAC51032.1 GBXI01014585 JAC99706.1 GAMC01021657 JAB84898.1 NEVH01018372 PNF23654.1 ACPB03005357 CP012528 ALC49049.1 CH940653 EDW62762.2 GL447388 EFN86491.1 GDHC01007920 JAQ10709.1 UFQT01000707 SSX26677.1 CH933810 EDW06653.1 GGFK01012271 MBW45592.1 KK854004 PTY09052.1 AY061462 AAF46213.2 AAL29010.1 LJIG01016162 KRT81428.1 PYGN01015190 PSN29002.1
KQ460761 KPJ12531.1 JTDY01001767 KOB72945.1 GFDL01011067 JAV23978.1 ADMH02000413 ETN66623.1 DS232478 EDS42365.1 GDUN01000184 JAN95735.1 GGFL01002678 MBW66856.1 APCN01003693 GEZM01033279 JAV84288.1 KQ971338 KYB27968.1 AXCN02000558 AXCM01000934 GBYB01007987 JAG77754.1 GBYB01013557 GBYB01013558 JAG83324.1 JAG83325.1 ATLV01019208 KE525264 KFB43936.1 KQ434869 KZC09378.1 KK107906 QOIP01000010 EZA47258.1 RLU17654.1 GFDF01010894 JAV03190.1 KQ414642 KOC66719.1 GL438568 EFN68816.1 GL769036 EFZ10812.1 GDAI01002887 JAI14716.1 KQ977858 KYM99313.1 CH477605 EAT38385.1 GL887707 EGI70239.1 KQ981523 KYN40413.1 AAAB01008807 EAA04358.3 GECZ01024567 GECZ01015766 JAS45202.1 JAS54003.1 NNAY01001413 OXU24056.1 KZ288449 PBC25594.1 KB631581 ERL84482.1 GECU01001175 JAT06532.1 KQ976731 KYM76262.1 DS235053 EEB11032.1 KQ983112 KYQ47356.1 ADTU01006966 KQ435951 KOX68113.1 APGK01031920 KB740831 ENN78817.1 KQ980724 KYN14082.1 GEBQ01004926 JAT35051.1 CM000162 EDX01060.1 CH954180 EDV47422.1 GECU01036773 JAS70933.1 CH964239 EDW82217.1 GBHO01022996 GBHO01022995 GBHO01022991 GBHO01022990 GBHO01001436 GBHO01001418 GDHC01015722 JAG20608.1 JAG20609.1 JAG20613.1 JAG20614.1 JAG42168.1 JAG42186.1 JAQ02907.1 CCAG010021577 EZ423281 ADD19557.1 OUUW01000003 SPP77796.1 JXJN01002588 CH902622 EDV34617.1 CH916371 EDV92223.1 AE014298 ACZ95219.1 GEDC01020592 JAS16706.1 GEDC01019592 JAS17706.1 CH379063 EAL32276.2 GDHF01033302 GDHF01006278 JAI19012.1 JAI46036.1 GAKP01007920 JAC51032.1 GBXI01014585 JAC99706.1 GAMC01021657 JAB84898.1 NEVH01018372 PNF23654.1 ACPB03005357 CP012528 ALC49049.1 CH940653 EDW62762.2 GL447388 EFN86491.1 GDHC01007920 JAQ10709.1 UFQT01000707 SSX26677.1 CH933810 EDW06653.1 GGFK01012271 MBW45592.1 KK854004 PTY09052.1 AY061462 AAF46213.2 AAL29010.1 LJIG01016162 KRT81428.1 PYGN01015190 PSN29002.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000037510
UP000000673
UP000002320
+ More
UP000075884 UP000075900 UP000075920 UP000075881 UP000076407 UP000075882 UP000075840 UP000075902 UP000069272 UP000007266 UP000075886 UP000075880 UP000075883 UP000075903 UP000030765 UP000192223 UP000076502 UP000053097 UP000279307 UP000053825 UP000000311 UP000078542 UP000008820 UP000075885 UP000007755 UP000078541 UP000007062 UP000215335 UP000242457 UP000030742 UP000078540 UP000009046 UP000075809 UP000005205 UP000005203 UP000053105 UP000019118 UP000078492 UP000002282 UP000008711 UP000007798 UP000092444 UP000092445 UP000192221 UP000078200 UP000268350 UP000091820 UP000092443 UP000092460 UP000007801 UP000001070 UP000000803 UP000001819 UP000076408 UP000235965 UP000015103 UP000095301 UP000095300 UP000092553 UP000008792 UP000008237 UP000009192 UP000245037
UP000075884 UP000075900 UP000075920 UP000075881 UP000076407 UP000075882 UP000075840 UP000075902 UP000069272 UP000007266 UP000075886 UP000075880 UP000075883 UP000075903 UP000030765 UP000192223 UP000076502 UP000053097 UP000279307 UP000053825 UP000000311 UP000078542 UP000008820 UP000075885 UP000007755 UP000078541 UP000007062 UP000215335 UP000242457 UP000030742 UP000078540 UP000009046 UP000075809 UP000005205 UP000005203 UP000053105 UP000019118 UP000078492 UP000002282 UP000008711 UP000007798 UP000092444 UP000092445 UP000192221 UP000078200 UP000268350 UP000091820 UP000092443 UP000092460 UP000007801 UP000001070 UP000000803 UP000001819 UP000076408 UP000235965 UP000015103 UP000095301 UP000095300 UP000092553 UP000008792 UP000008237 UP000009192 UP000245037
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A2A4J4K0
A0A2H1V440
A0A212FNX6
A0A194R9J4
A0A0L7LC29
A0A1Q3F904
+ More
W5JTL5 B0X896 A0A182N9A7 A0A182RJL0 A0A0P6JSP8 A0A1S4FNA9 A0A182WBI3 A0A182JRN7 A0A2M4CNF8 A0A182WWI1 A0A182L3N6 A0A182I6R1 A0A182TZD4 A0A182FEM3 A0A1Y1MH36 A0A139WIX5 A0A182Q8N8 A0A182IQX4 A0A182LTG1 A0A182VAG1 A0A0C9R5D3 A0A0C9RJU9 A0A084W142 A0A1W4XRP4 A0A154PDQ1 A0A026VTY1 A0A1L8DA30 A0A0L7R7A2 E2ACS2 E9J8M0 A0A0K8TKR0 A0A151IES1 Q16V43 A0A182PIJ2 F4W6A4 A0A195FJ32 Q7QIL4 A0A1B6FVE1 A0A232EZW6 A0A2A3E237 U4TUN5 A0A1B6K505 A0A195AW28 E0VCC6 A0A151WHP1 A0A158P220 A0A088A2R4 A0A0M8ZRF4 N6UJH6 A0A195DMM2 A0A1B6MGP5 B4PZ93 B3NXX1 A0A1B6H8I9 B4NEP8 A0A0A9ZFG2 D3TPC9 A0A1B0ABV9 A0A1W4V9R3 A0A1A9UHM6 A0A3B0JWR5 A0A1A9W1W6 A0A1A9XIB8 A0A1B0ARV5 B3MS62 B4JND0 E1JJF2 A0A1B6CT62 A0A1B6CW23 Q29JM5 A0A0K8W4K8 A0A034W9P2 A0A182Y2B6 A0A0A1WLH7 W8AJM0 A0A2J7Q502 T1IB28 A0A1I8NC67 A0A1I8Q8X2 A0A0M3QZA8 B4M7J5 E2BCT9 A0A146LTY3 A0A336ME62 B4L1D2 A0A2M4AY05 A0A2R7VNB8 Q9W3V0 A0A0T6B2F1 A0A2P8XAH9
W5JTL5 B0X896 A0A182N9A7 A0A182RJL0 A0A0P6JSP8 A0A1S4FNA9 A0A182WBI3 A0A182JRN7 A0A2M4CNF8 A0A182WWI1 A0A182L3N6 A0A182I6R1 A0A182TZD4 A0A182FEM3 A0A1Y1MH36 A0A139WIX5 A0A182Q8N8 A0A182IQX4 A0A182LTG1 A0A182VAG1 A0A0C9R5D3 A0A0C9RJU9 A0A084W142 A0A1W4XRP4 A0A154PDQ1 A0A026VTY1 A0A1L8DA30 A0A0L7R7A2 E2ACS2 E9J8M0 A0A0K8TKR0 A0A151IES1 Q16V43 A0A182PIJ2 F4W6A4 A0A195FJ32 Q7QIL4 A0A1B6FVE1 A0A232EZW6 A0A2A3E237 U4TUN5 A0A1B6K505 A0A195AW28 E0VCC6 A0A151WHP1 A0A158P220 A0A088A2R4 A0A0M8ZRF4 N6UJH6 A0A195DMM2 A0A1B6MGP5 B4PZ93 B3NXX1 A0A1B6H8I9 B4NEP8 A0A0A9ZFG2 D3TPC9 A0A1B0ABV9 A0A1W4V9R3 A0A1A9UHM6 A0A3B0JWR5 A0A1A9W1W6 A0A1A9XIB8 A0A1B0ARV5 B3MS62 B4JND0 E1JJF2 A0A1B6CT62 A0A1B6CW23 Q29JM5 A0A0K8W4K8 A0A034W9P2 A0A182Y2B6 A0A0A1WLH7 W8AJM0 A0A2J7Q502 T1IB28 A0A1I8NC67 A0A1I8Q8X2 A0A0M3QZA8 B4M7J5 E2BCT9 A0A146LTY3 A0A336ME62 B4L1D2 A0A2M4AY05 A0A2R7VNB8 Q9W3V0 A0A0T6B2F1 A0A2P8XAH9
Ontologies
PANTHER
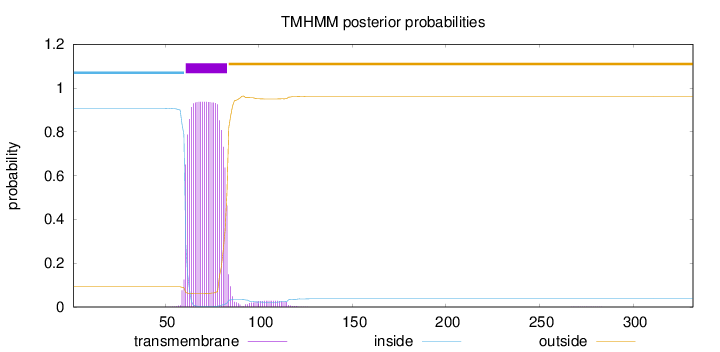
Topology
Length:
332
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.97159
Exp number, first 60 AAs:
0.22325
Total prob of N-in:
0.90736
inside
1 - 60
TMhelix
61 - 83
outside
84 - 332
Population Genetic Test Statistics
Pi
197.722963
Theta
176.913023
Tajima's D
0.314335
CLR
0.090306
CSRT
0.460376981150942
Interpretation
Uncertain
