Gene
KWMTBOMO07987
Annotation
PREDICTED:_probable_endochitinase_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.674 PlasmaMembrane Reliability : 1.223
Sequence
CDS
ATGCGTTTTGTAAATTTTATTGAGAAGGCAAAAATGAAACGAAAGGCTGGCATAGTTTTAATACTCGTTTTCTGTTGCGTTTGGCAAGTTCAGGCAGCCGTCAACTGTACGGAGACTGGAGCAGGCAGATTTGCCGATGACGATGACACGACTTGTCAAAACTACACGCTTTGCGTCTACGATACTACGACTTTAGATTATTTGTTCTACAACTATCAATGTCCATCGACGTCTGTGTTCAATCCGAACATTGCAATGTGTACCGATCCTGCCAACTACGTATGCAATGTGACGACCGCAAATACTACCGTTTGTACCGAAGACGGTCTCACTGACATGTCGAGTTTCATTATTGTCGTTAAAATCCGGAACAGCGATGACCAGGTACGCGCTTTCATTAATTTGATGAACTTGATACGTTTTAAAACAAAACGTTAG
Protein
MRFVNFIEKAKMKRKAGIVLILVFCCVWQVQAAVNCTETGAGRFADDDDTTCQNYTLCVYDTTTLDYLFYNYQCPSTSVFNPNIAMCTDPANYVCNVTTANTTVCTEDGLTDMSSFIIVVKIRNSDDQVRAFINLMNLIRFKTKR
Summary
Uniprot
EMBL
Proteomes
Pfam
PF01607 CBM_14
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
Ontologies
GO
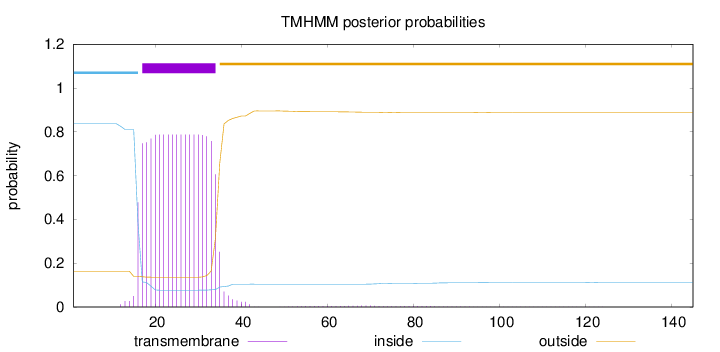
Topology
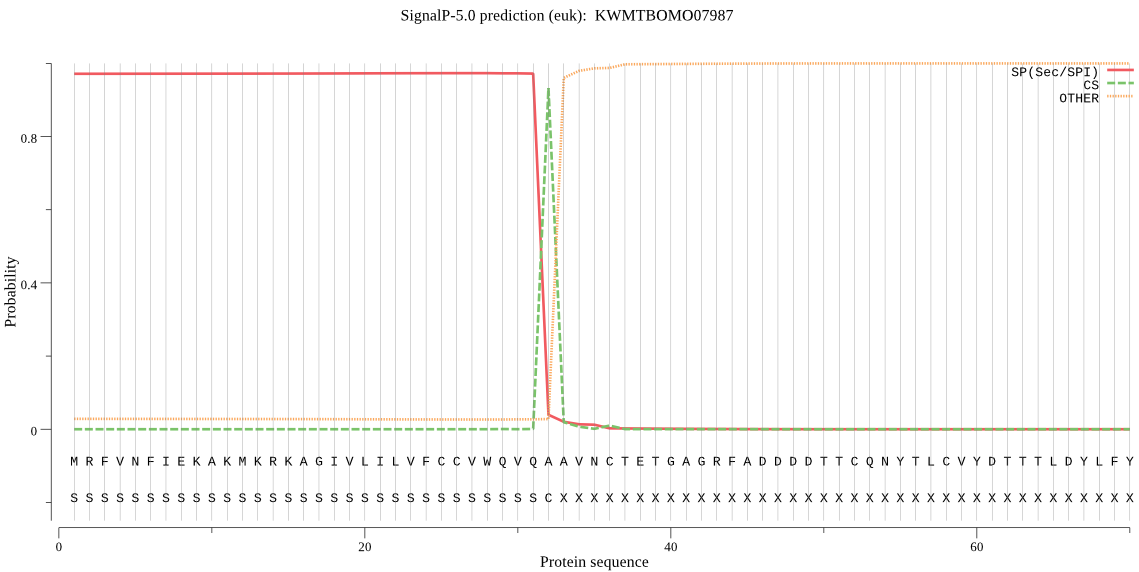
SignalP
Position: 1 - 32,
Likelihood: 0.972930
Length:
145
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
15.12606
Exp number, first 60 AAs:
14.98489
Total prob of N-in:
0.83678
POSSIBLE N-term signal
sequence
inside
1 - 16
TMhelix
17 - 34
outside
35 - 145
Population Genetic Test Statistics
Pi
153.072877
Theta
159.031513
Tajima's D
0.040338
CLR
0
CSRT
0.384280785960702
Interpretation
Uncertain
