Gene
KWMTBOMO07981
Pre Gene Modal
BGIBMGA001145
Annotation
PREDICTED:_uncharacterized_protein_LOC101742398_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.862
Sequence
CDS
ATGGATACCCGGTCGAAAGGGGTAAGGGACTGGGTTATGTCGGACTCCGGCACCTCCAGAGAGGAAGAGGGAGACGAGGAGGGCCAAGGTAAAAGGATTTTATCTTGCTTTTCGACGAGATCTCTGGAGCCGTCTTTCGGTCTCCTATTCAAAGACTTGTTGGATGACCTTGGCGTCCGCACAACGGGTGCTTCCTTCATCGCTAGTACTTTGGACTCTGTAGTAAATTTCTCCGGCTTCTTCGTTGGTCCTGTTATAAAGACGTTCTCATACAGAAAAGTTTGTTTCGTCGGTTCAACGATATGTGCCCTCGGATTACTGTTCACCGCCCCGGCGAAGAGTATCGGCCATATATTGGCGACCTACAGCATTTTGGGAGGGTTTGGCGTAGGTCTAGCCTCGTCTTCTTCTTTTGTCTCCCTCAATCACTATTTCTCCAAGAAGCGAGGACAGGCCGTGGGTTTGTCCATGGCGGGAACCGGGTTCGGTTTGATGGTGATGCCACAACTAGTCAAATTACTACTGGGAGAGTACGGGTTCAGATGGACCGTAGTGATACTTGGAGCGCTGGCGTTTAACGCTGTAGTAGGATCATGTCTCCTTCAACCAATCAAAAGGCATCTAAAGGAAGTACCCGTGGATGCGGAAATGCAAAACATTAAAGAGATGGAATGCATTATGGAAAGCGACGAAGAAAACGAAGATAAATTCGAGATCAATCCTCTCGTCACGCATCCGATGCCGAAACTAACAACGCAGCGTTCCCACTCTAACATGAATCATCCTTCGGTTCGCACGATAGGGATGACCCGCGCTAAGACCTGCGATAAACCGTTACAAGATTTTGAGAAGAACTCGAAGGTGTACCCGGGGCTCACCACCGCGGCCTCTGTGGGCGTGATGCCGCGAGTGACATCTAGCGGCAGCATGAACGAAGCGGCACGCAGAAGGAAAGTCTCCGTCATCTCCAACATATCAAACATGGACTTCACGGGCAGCTATCTACAGCTCTATCTCGATACAGCAGACGAAGAAGCGATACAAATGAAGACAATAAAAAAAGCCGAGCCCGTATCGGAAAAGAAGGGCTTCATTAGACGATTCATAGATCTGATGGATTTAGATTTGCTAAAGGACTGGTCCTTTCTGAACCTAATCCTAGGACTGGCTCTGTTTTGGAGCGGCGAGTTGCAGTTCAGGATGTTGACCCCGTTCTTTATAAGCAATTTAGGTTACAACATGGACGACACGGCGTTCTGTCTGTCGATGACCGCGATCTCCGATAACCTGGTGCGGCTGATCCTACCACCGATTTTCGACAGAACCACGATATCGAAGAAATTGATATTTTTTATATCCTCGTTTTTCTTGGCCGCCACTAGATCAGTACTAGCCGAGCAAACTGAATGGGTGTCGCTGATGATATGGCTGTCTATATGCGGTTTCTTCCGTGGGATGTGTCTCAGCAACTTCACCCTAACGATATCCGAGTACTGTCCCTTGGAGAAACTGCCCGCGGCTTTCGGACTGCACATGGTCAGCAAGGGTGTGCTCGTCGTATTGATCGGGCCAATGATTGGATACGTAAGGGATTACACGGGCAGTTTCGCTCTATGCATTCACGTGCAAAACGCAATGATAATGTCCTGCGTTTTAGTTTGGGGCATCGAATACGTTGTAACGTTAACGAGACGCAGGAAGATCGTCCAAATATAA
Protein
MDTRSKGVRDWVMSDSGTSREEEGDEEGQGKRILSCFSTRSLEPSFGLLFKDLLDDLGVRTTGASFIASTLDSVVNFSGFFVGPVIKTFSYRKVCFVGSTICALGLLFTAPAKSIGHILATYSILGGFGVGLASSSSFVSLNHYFSKKRGQAVGLSMAGTGFGLMVMPQLVKLLLGEYGFRWTVVILGALAFNAVVGSCLLQPIKRHLKEVPVDAEMQNIKEMECIMESDEENEDKFEINPLVTHPMPKLTTQRSHSNMNHPSVRTIGMTRAKTCDKPLQDFEKNSKVYPGLTTAASVGVMPRVTSSGSMNEAARRRKVSVISNISNMDFTGSYLQLYLDTADEEAIQMKTIKKAEPVSEKKGFIRRFIDLMDLDLLKDWSFLNLILGLALFWSGELQFRMLTPFFISNLGYNMDDTAFCLSMTAISDNLVRLILPPIFDRTTISKKLIFFISSFFLAATRSVLAEQTEWVSLMIWLSICGFFRGMCLSNFTLTISEYCPLEKLPAAFGLHMVSKGVLVVLIGPMIGYVRDYTGSFALCIHVQNAMIMSCVLVWGIEYVVTLTRRRKIVQI
Summary
Uniprot
H9IV65
A0A2H1V1E0
A0A212FP05
A0A2A4JNS2
A0A194R402
A0A1A9W689
+ More
A0A1B0FC82 A0A1A9ZB17 A0A1A9UZI6 A0A0A1XFI4 A0A0K8VW88 A0A034V7K4 W8AE90 A0A1I8PQM0 T1PG59 B4M2A1 A0A1B0C7R1 A0A1A9YMB2 B4I742 B4L851 B3NVM6 B4PZJ3 Q9VWJ1 A0A1I8PQH5 A0A1B0DB11 B3N141 A0A1W4VFY9 A0A3B0KPW6 B4GVE0 A0A0M4EUF5 B4JK09 B4MT02 A0A1L8DEN3 A0A2J7RLJ5 Q29GP5 A0A1J1I0N8 Q16Z20 A0A1Q3G203 A0A1Q3G1R7 Q7K0I7 A0A084VVS1 A0A023EU72 A0A182GUD2 D2A1Q6 A0A182MC36 A0A182QNB6 A0A182TBY0 A0A182W4B8 A0A182Y7K4 A0A182K457 V5GRR8 A0A0T6AZG7 A0A182R3H0 A0A182I9R6 A0A182PPC2 A0A182V6W5 A0A182X570 Q7QEC4 A0A182IQF0 A0A1L8DEG8 A0A182FJZ5 A0A2M3Z4P2 A0A2M3Z4I7 A0A182TIQ2 A0A1Q3G1Y0 A0A1Q3G1W0 A0A182MZR4 A0A3B0KJD2 A0A2J7RLK3 A0A2J7RLK9 A0A2J7RLJ6 A0A1Y1LYE8 A0A1B6C167 U4UMS7 A0A2P8YB28 A0A336M8U9 W5JJ46 A0A182L6A8 A0A0K8SYG5 A0A023F750 A0A0V0G6D4 A0A2R7X361 A0A224XLU6 R4G8Y9 A0A2J7RLK2 A0A1L8DDT6 A0A310SHY0 A0A0L7R3M1 A0A1B6LCF0 A0A2P8YB42 N6TDX2 A0A1I8MT93
A0A1B0FC82 A0A1A9ZB17 A0A1A9UZI6 A0A0A1XFI4 A0A0K8VW88 A0A034V7K4 W8AE90 A0A1I8PQM0 T1PG59 B4M2A1 A0A1B0C7R1 A0A1A9YMB2 B4I742 B4L851 B3NVM6 B4PZJ3 Q9VWJ1 A0A1I8PQH5 A0A1B0DB11 B3N141 A0A1W4VFY9 A0A3B0KPW6 B4GVE0 A0A0M4EUF5 B4JK09 B4MT02 A0A1L8DEN3 A0A2J7RLJ5 Q29GP5 A0A1J1I0N8 Q16Z20 A0A1Q3G203 A0A1Q3G1R7 Q7K0I7 A0A084VVS1 A0A023EU72 A0A182GUD2 D2A1Q6 A0A182MC36 A0A182QNB6 A0A182TBY0 A0A182W4B8 A0A182Y7K4 A0A182K457 V5GRR8 A0A0T6AZG7 A0A182R3H0 A0A182I9R6 A0A182PPC2 A0A182V6W5 A0A182X570 Q7QEC4 A0A182IQF0 A0A1L8DEG8 A0A182FJZ5 A0A2M3Z4P2 A0A2M3Z4I7 A0A182TIQ2 A0A1Q3G1Y0 A0A1Q3G1W0 A0A182MZR4 A0A3B0KJD2 A0A2J7RLK3 A0A2J7RLK9 A0A2J7RLJ6 A0A1Y1LYE8 A0A1B6C167 U4UMS7 A0A2P8YB28 A0A336M8U9 W5JJ46 A0A182L6A8 A0A0K8SYG5 A0A023F750 A0A0V0G6D4 A0A2R7X361 A0A224XLU6 R4G8Y9 A0A2J7RLK2 A0A1L8DDT6 A0A310SHY0 A0A0L7R3M1 A0A1B6LCF0 A0A2P8YB42 N6TDX2 A0A1I8MT93
Pubmed
19121390
22118469
26354079
25830018
25348373
24495485
+ More
25315136 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17510324 26109357 26109356 24438588 24945155 26483478 18362917 19820115 25244985 12364791 14747013 17210077 28004739 23537049 29403074 20920257 23761445 20966253 25474469
25315136 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17510324 26109357 26109356 24438588 24945155 26483478 18362917 19820115 25244985 12364791 14747013 17210077 28004739 23537049 29403074 20920257 23761445 20966253 25474469
EMBL
BABH01000572
ODYU01000236
SOQ34667.1
AGBW02004200
OWR55439.1
NWSH01001018
+ More
PCG73093.1 KQ460761 KPJ12538.1 CCAG010003149 GBXI01012470 GBXI01009867 GBXI01005049 GBXI01001190 JAD01822.1 JAD04425.1 JAD09243.1 JAD13102.1 GDHF01009166 JAI43148.1 GAKP01019693 GAKP01019691 JAC39261.1 GAMC01020085 GAMC01020084 JAB86470.1 KA647669 AFP62298.1 CH940651 EDW65805.1 JXJN01030313 CH480823 EDW56140.1 CH933814 EDW05626.1 KRG07465.1 KRG07466.1 CH954180 EDV46691.1 CM000162 EDX02149.1 AE014298 AAF48947.2 AJVK01000597 AJVK01000598 AJVK01000599 CH902650 EDV30076.1 OUUW01000015 SPP88659.1 CH479192 EDW26427.1 CP012528 ALC48710.1 CH916370 EDV99911.1 CH963851 EDW75241.1 GFDF01009161 JAV04923.1 NEVH01002684 PNF41714.1 CH379064 EAL32065.3 CVRI01000036 CRK93148.1 CH477503 EAT39889.1 GFDL01001222 JAV33823.1 GFDL01001303 JAV33742.1 AY069772 AAL39917.1 ACZ95328.1 ATLV01017216 KE525157 KFB42065.1 GAPW01000648 JAC12950.1 JXUM01088920 JXUM01088921 JXUM01088922 JXUM01088923 JXUM01088924 JXUM01088925 JXUM01088926 KQ971338 EFA02707.1 AXCM01002930 AXCN02001217 GALX01004139 JAB64327.1 LJIG01022462 KRT80450.1 APCN01001029 AAAB01008847 EAA06880.5 GFDF01009309 JAV04775.1 GGFM01002667 MBW23418.1 GGFM01002675 MBW23426.1 GFDL01001231 JAV33814.1 GFDL01001299 JAV33746.1 SPP88660.1 PNF41712.1 PNF41715.1 PNF41716.1 GEZM01043968 GEZM01043967 GEZM01043966 JAV78624.1 GEDC01030051 JAS07247.1 KB632374 ERL93798.1 PYGN01000741 PSN41451.1 UFQT01000677 SSX26480.1 ADMH02001354 ETN62895.1 GBRD01007615 GBRD01003957 JAG58206.1 GBBI01001501 JAC17211.1 GECL01002473 JAP03651.1 KK856674 PTY26239.1 GFTR01007016 JAW09410.1 GAHY01000142 JAA77368.1 PNF41710.1 GFDF01009476 JAV04608.1 KQ761046 OAD58323.1 KQ414663 KOC65429.1 GEBQ01018599 JAT21378.1 PSN41453.1 APGK01033162 APGK01033163 KB740862 ENN78554.1
PCG73093.1 KQ460761 KPJ12538.1 CCAG010003149 GBXI01012470 GBXI01009867 GBXI01005049 GBXI01001190 JAD01822.1 JAD04425.1 JAD09243.1 JAD13102.1 GDHF01009166 JAI43148.1 GAKP01019693 GAKP01019691 JAC39261.1 GAMC01020085 GAMC01020084 JAB86470.1 KA647669 AFP62298.1 CH940651 EDW65805.1 JXJN01030313 CH480823 EDW56140.1 CH933814 EDW05626.1 KRG07465.1 KRG07466.1 CH954180 EDV46691.1 CM000162 EDX02149.1 AE014298 AAF48947.2 AJVK01000597 AJVK01000598 AJVK01000599 CH902650 EDV30076.1 OUUW01000015 SPP88659.1 CH479192 EDW26427.1 CP012528 ALC48710.1 CH916370 EDV99911.1 CH963851 EDW75241.1 GFDF01009161 JAV04923.1 NEVH01002684 PNF41714.1 CH379064 EAL32065.3 CVRI01000036 CRK93148.1 CH477503 EAT39889.1 GFDL01001222 JAV33823.1 GFDL01001303 JAV33742.1 AY069772 AAL39917.1 ACZ95328.1 ATLV01017216 KE525157 KFB42065.1 GAPW01000648 JAC12950.1 JXUM01088920 JXUM01088921 JXUM01088922 JXUM01088923 JXUM01088924 JXUM01088925 JXUM01088926 KQ971338 EFA02707.1 AXCM01002930 AXCN02001217 GALX01004139 JAB64327.1 LJIG01022462 KRT80450.1 APCN01001029 AAAB01008847 EAA06880.5 GFDF01009309 JAV04775.1 GGFM01002667 MBW23418.1 GGFM01002675 MBW23426.1 GFDL01001231 JAV33814.1 GFDL01001299 JAV33746.1 SPP88660.1 PNF41712.1 PNF41715.1 PNF41716.1 GEZM01043968 GEZM01043967 GEZM01043966 JAV78624.1 GEDC01030051 JAS07247.1 KB632374 ERL93798.1 PYGN01000741 PSN41451.1 UFQT01000677 SSX26480.1 ADMH02001354 ETN62895.1 GBRD01007615 GBRD01003957 JAG58206.1 GBBI01001501 JAC17211.1 GECL01002473 JAP03651.1 KK856674 PTY26239.1 GFTR01007016 JAW09410.1 GAHY01000142 JAA77368.1 PNF41710.1 GFDF01009476 JAV04608.1 KQ761046 OAD58323.1 KQ414663 KOC65429.1 GEBQ01018599 JAT21378.1 PSN41453.1 APGK01033162 APGK01033163 KB740862 ENN78554.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000091820
UP000092444
+ More
UP000092445 UP000078200 UP000095300 UP000095301 UP000008792 UP000092460 UP000092443 UP000001292 UP000009192 UP000008711 UP000002282 UP000000803 UP000092462 UP000007801 UP000192221 UP000268350 UP000008744 UP000092553 UP000001070 UP000007798 UP000235965 UP000001819 UP000183832 UP000008820 UP000030765 UP000069940 UP000007266 UP000075883 UP000075886 UP000075901 UP000075920 UP000076408 UP000075881 UP000075900 UP000075840 UP000075885 UP000075903 UP000076407 UP000007062 UP000075880 UP000069272 UP000075902 UP000075884 UP000030742 UP000245037 UP000000673 UP000075882 UP000053825 UP000019118
UP000092445 UP000078200 UP000095300 UP000095301 UP000008792 UP000092460 UP000092443 UP000001292 UP000009192 UP000008711 UP000002282 UP000000803 UP000092462 UP000007801 UP000192221 UP000268350 UP000008744 UP000092553 UP000001070 UP000007798 UP000235965 UP000001819 UP000183832 UP000008820 UP000030765 UP000069940 UP000007266 UP000075883 UP000075886 UP000075901 UP000075920 UP000076408 UP000075881 UP000075900 UP000075840 UP000075885 UP000075903 UP000076407 UP000007062 UP000075880 UP000069272 UP000075902 UP000075884 UP000030742 UP000245037 UP000000673 UP000075882 UP000053825 UP000019118
Pfam
PF07690 MFS_1
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9IV65
A0A2H1V1E0
A0A212FP05
A0A2A4JNS2
A0A194R402
A0A1A9W689
+ More
A0A1B0FC82 A0A1A9ZB17 A0A1A9UZI6 A0A0A1XFI4 A0A0K8VW88 A0A034V7K4 W8AE90 A0A1I8PQM0 T1PG59 B4M2A1 A0A1B0C7R1 A0A1A9YMB2 B4I742 B4L851 B3NVM6 B4PZJ3 Q9VWJ1 A0A1I8PQH5 A0A1B0DB11 B3N141 A0A1W4VFY9 A0A3B0KPW6 B4GVE0 A0A0M4EUF5 B4JK09 B4MT02 A0A1L8DEN3 A0A2J7RLJ5 Q29GP5 A0A1J1I0N8 Q16Z20 A0A1Q3G203 A0A1Q3G1R7 Q7K0I7 A0A084VVS1 A0A023EU72 A0A182GUD2 D2A1Q6 A0A182MC36 A0A182QNB6 A0A182TBY0 A0A182W4B8 A0A182Y7K4 A0A182K457 V5GRR8 A0A0T6AZG7 A0A182R3H0 A0A182I9R6 A0A182PPC2 A0A182V6W5 A0A182X570 Q7QEC4 A0A182IQF0 A0A1L8DEG8 A0A182FJZ5 A0A2M3Z4P2 A0A2M3Z4I7 A0A182TIQ2 A0A1Q3G1Y0 A0A1Q3G1W0 A0A182MZR4 A0A3B0KJD2 A0A2J7RLK3 A0A2J7RLK9 A0A2J7RLJ6 A0A1Y1LYE8 A0A1B6C167 U4UMS7 A0A2P8YB28 A0A336M8U9 W5JJ46 A0A182L6A8 A0A0K8SYG5 A0A023F750 A0A0V0G6D4 A0A2R7X361 A0A224XLU6 R4G8Y9 A0A2J7RLK2 A0A1L8DDT6 A0A310SHY0 A0A0L7R3M1 A0A1B6LCF0 A0A2P8YB42 N6TDX2 A0A1I8MT93
A0A1B0FC82 A0A1A9ZB17 A0A1A9UZI6 A0A0A1XFI4 A0A0K8VW88 A0A034V7K4 W8AE90 A0A1I8PQM0 T1PG59 B4M2A1 A0A1B0C7R1 A0A1A9YMB2 B4I742 B4L851 B3NVM6 B4PZJ3 Q9VWJ1 A0A1I8PQH5 A0A1B0DB11 B3N141 A0A1W4VFY9 A0A3B0KPW6 B4GVE0 A0A0M4EUF5 B4JK09 B4MT02 A0A1L8DEN3 A0A2J7RLJ5 Q29GP5 A0A1J1I0N8 Q16Z20 A0A1Q3G203 A0A1Q3G1R7 Q7K0I7 A0A084VVS1 A0A023EU72 A0A182GUD2 D2A1Q6 A0A182MC36 A0A182QNB6 A0A182TBY0 A0A182W4B8 A0A182Y7K4 A0A182K457 V5GRR8 A0A0T6AZG7 A0A182R3H0 A0A182I9R6 A0A182PPC2 A0A182V6W5 A0A182X570 Q7QEC4 A0A182IQF0 A0A1L8DEG8 A0A182FJZ5 A0A2M3Z4P2 A0A2M3Z4I7 A0A182TIQ2 A0A1Q3G1Y0 A0A1Q3G1W0 A0A182MZR4 A0A3B0KJD2 A0A2J7RLK3 A0A2J7RLK9 A0A2J7RLJ6 A0A1Y1LYE8 A0A1B6C167 U4UMS7 A0A2P8YB28 A0A336M8U9 W5JJ46 A0A182L6A8 A0A0K8SYG5 A0A023F750 A0A0V0G6D4 A0A2R7X361 A0A224XLU6 R4G8Y9 A0A2J7RLK2 A0A1L8DDT6 A0A310SHY0 A0A0L7R3M1 A0A1B6LCF0 A0A2P8YB42 N6TDX2 A0A1I8MT93
Ontologies
GO
Topology
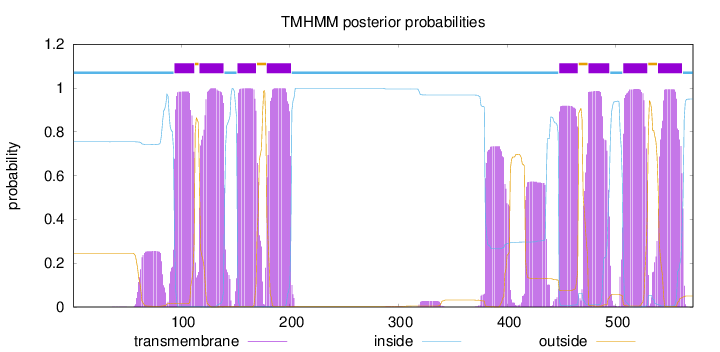
Length:
571
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
199.13932
Exp number, first 60 AAs:
0.12861
Total prob of N-in:
0.75447
inside
1 - 93
TMhelix
94 - 112
outside
113 - 116
TMhelix
117 - 139
inside
140 - 151
TMhelix
152 - 169
outside
170 - 178
TMhelix
179 - 201
inside
202 - 447
TMhelix
448 - 465
outside
466 - 474
TMhelix
475 - 494
inside
495 - 506
TMhelix
507 - 529
outside
530 - 538
TMhelix
539 - 561
inside
562 - 571
Population Genetic Test Statistics
Pi
213.855783
Theta
171.395398
Tajima's D
0.96682
CLR
0.106466
CSRT
0.648867556622169
Interpretation
Uncertain
