Gene
KWMTBOMO07978
Pre Gene Modal
BGIBMGA001003
Annotation
cytochrome_P450_4G49_[Manduca_sexta]
Location in the cell
Mitochondrial Reliability : 1.627 Nuclear Reliability : 1.016
Sequence
CDS
ATGATAATGTGGATGTTACACCGGTGGCAACAATCTTCTAGGCTGTTTCGATTGGGAAATAAACTCCCTGGACCAATGGCTTTACCTTTAGTGGGTAATTCGCTACTGATTTTAGGGAAAAAAGCTGAAGATTTAGTAAAATATGCTCTGGATTACTCTGAAAAATATGGAACTGTTGTGCGTGCCTGGGCTGGCCCAAAGCTCGTCGTGTTTCTAACAGACGCTAACGATGTCGAAGTAATTCTTAACAGTCAAATTCACATCGACAAGTCTCCCGAATATAGATTTTTCAAACCGTGGCTTGGAGAAGGATTACTTATCAGTTCAGGTGAAAAATGGCGTTCACATCGAAAAATGATCGCTCCGACATTCCATATAAATATTTTGAAATCGTTCATGGGAGTTTTCAATGAGAACAGCAAGAGCGTTGTTAAAAAATTACGTTCTGAAGTTGGAAAAACATTTGACGTTCACGATTACATGAGCTGCGTGACAGTCGATATCCTACTAGAGACAGCAATGGGAATAACAAAAACGACGCAAGATGCTGCGAGTTTTGACTATGCTATGGCCGTAATGAAAATGTGCAATATAATTCATCAGAGGCACTACAAAGTTTGGTTACATTTTGATGCCATATTCAAATTGACTTCACTTTTCAAAAAACAGAGGGAGCTTTTGAAAACAATTCATGGTTTAACAAATAAGGTTATAAAAAAAAAGAAATTCATGTACCTACAAAACAAAGAAAAAGGCATAATTCCACCAACAATAGAAGAACTGACTAAAATAAAAGATACAAATGACAGCATAATGGAAGATTCTGCAAAAACACTTTCCGATACAGTTTTTAAGGGCTATCGTGATGATTTGGATTTTAATGATGAACAAGATGTTGGCGAAAAGAAGCGTCTCGCTTTCTTAGACTTGATGATTGAATCTGCACAAAACCACACATGTAATATCAGCGACCACGAAATCAAAGAGGAAGTTGACACTATTATGTTTGAGGGACATGACACCACAGCAGCTGGATCAAGCTTTGTCCTCTGTTTGTTGGGTATTCATCAAGAAATTCAGAGTAAAGTATATGATGAATTATTTGAAATCTTCGGAGATTCTGACAGACTGGTGACATTTGCTGACACGCTTCAAATGAAGTACTTAGAGAGGGTAATTCTGGAGTCTCTCAGATTGTATCCACCAGTGCCGGCTATAGCAAGGAAGCTGACGCGAGATGTCCAGATCGTTACAAACAACTACATTATACCAGCGGGAAGTACCGTCGTTATTGGAACGTTCAAAATTCATCGTGATCCTAAATATCATAAGAACCCGAATGTTTTCAATCCAGACAACTTCCTGCCAGAAAACACACAAAATAGGCACTACTACAGCTACATACCGTTCAGCGCTGGGCCAAGAAGCTGTGTCGGTCGCAAATATGCTTTACTTAAACTGAAAGTGCTGCTCTCGACAATTCTTCGTAATTACAAAACCACATCCGAAATATCTGAAGATCAATTTGTACTTCAGGCAGACATCATTTTGAAGAGATACGATGGCTTCAAAATTCGAATTGAACCAAGGAATAAAAATCATTCAAATACTGTTTAA
Protein
MIMWMLHRWQQSSRLFRLGNKLPGPMALPLVGNSLLILGKKAEDLVKYALDYSEKYGTVVRAWAGPKLVVFLTDANDVEVILNSQIHIDKSPEYRFFKPWLGEGLLISSGEKWRSHRKMIAPTFHINILKSFMGVFNENSKSVVKKLRSEVGKTFDVHDYMSCVTVDILLETAMGITKTTQDAASFDYAMAVMKMCNIIHQRHYKVWLHFDAIFKLTSLFKKQRELLKTIHGLTNKVIKKKKFMYLQNKEKGIIPPTIEELTKIKDTNDSIMEDSAKTLSDTVFKGYRDDLDFNDEQDVGEKKRLAFLDLMIESAQNHTCNISDHEIKEEVDTIMFEGHDTTAAGSSFVLCLLGIHQEIQSKVYDELFEIFGDSDRLVTFADTLQMKYLERVILESLRLYPPVPAIARKLTRDVQIVTNNYIIPAGSTVVIGTFKIHRDPKYHKNPNVFNPDNFLPENTQNRHYYSYIPFSAGPRSCVGRKYALLKLKVLLSTILRNYKTTSEISEDQFVLQADIILKRYDGFKIRIEPRNKNHSNTV
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
L0N4R3
D2JLK4
D5L0M8
H9IUS3
A0A2A4JNU7
A0A068F0V7
+ More
S4VAQ8 A0A248QEH6 A0A1V0D9D0 A0A194QA50 A0A2A4JNR2 L0N788 L0N4H7 L0N7D2 A0A2A4JM80 Q1HPP0 A0A0C5C1N9 A0A068ETT4 A0A2H1V7J5 A0A3S2TKW7 H9IUS4 A0A194R5M7 A0A3S2P026 X5DB06 A0A0L7L7K6 L0N5J9 A0A1L8E432 X5D4H8 Q174P3 A0A1S4FEN8 A0A1Q3FRN6 Q16Z24 A0A1Q3FR81 A0A1Q3FRH5 A0A1Q3FRH0 A0A182GGE4 B0XHB7 K7J4F5 A0A232FC79 A0A2S0E492 A0A2A4JMP1 A0A182RYV2 A0A182UQZ2 A0A182U8B6 A0A182KW88 Q5TQQ1 A0A182HMS7 A0A182WY36 A0A182MQZ0 A0A182FJZ7 A0A182Q0J4 A0A182J321 A0A182TCR7 A0A0B5HSJ6 A0A084VVS6 A0A182W664 A0A182XYA1 A0A182P0M1 A0A182KG08 A0A1B0GJR2 A0A182MZX3 W5JVS3 A0A0C9R9S5 F5HL83 A0A182IDW2 A0A182VA75 A0A1J1HSQ0 V5T6Q9 Q3I413 B0WQV0 A0A1Q3FQS4 A0A2J7RLH7 A0A0L0CJW1 A0A1L1V8K0 A0A1Q3FR66 A0A0K1YWB8 A0A2H1VJ39 A0A125S6P4 A0A1J1I034 A0A182LRX0 A0A0K8UW61 A0A182W5Z8 D1ZZJ8 A0A084VQ44 T1E750 A0A182IJ93 A0A182YH39 A0A1B1W254 A0A0A1X7F2 A0A182RVW8 Q5EI69 A0A1S3D5P4 A0A2M3Z3M6 A0A195F9G5 A0A182N9Y9 D5L0M7 A0A182PU94 A0A2M3Z3F0 W5JIT2 A0A1W4V117
S4VAQ8 A0A248QEH6 A0A1V0D9D0 A0A194QA50 A0A2A4JNR2 L0N788 L0N4H7 L0N7D2 A0A2A4JM80 Q1HPP0 A0A0C5C1N9 A0A068ETT4 A0A2H1V7J5 A0A3S2TKW7 H9IUS4 A0A194R5M7 A0A3S2P026 X5DB06 A0A0L7L7K6 L0N5J9 A0A1L8E432 X5D4H8 Q174P3 A0A1S4FEN8 A0A1Q3FRN6 Q16Z24 A0A1Q3FR81 A0A1Q3FRH5 A0A1Q3FRH0 A0A182GGE4 B0XHB7 K7J4F5 A0A232FC79 A0A2S0E492 A0A2A4JMP1 A0A182RYV2 A0A182UQZ2 A0A182U8B6 A0A182KW88 Q5TQQ1 A0A182HMS7 A0A182WY36 A0A182MQZ0 A0A182FJZ7 A0A182Q0J4 A0A182J321 A0A182TCR7 A0A0B5HSJ6 A0A084VVS6 A0A182W664 A0A182XYA1 A0A182P0M1 A0A182KG08 A0A1B0GJR2 A0A182MZX3 W5JVS3 A0A0C9R9S5 F5HL83 A0A182IDW2 A0A182VA75 A0A1J1HSQ0 V5T6Q9 Q3I413 B0WQV0 A0A1Q3FQS4 A0A2J7RLH7 A0A0L0CJW1 A0A1L1V8K0 A0A1Q3FR66 A0A0K1YWB8 A0A2H1VJ39 A0A125S6P4 A0A1J1I034 A0A182LRX0 A0A0K8UW61 A0A182W5Z8 D1ZZJ8 A0A084VQ44 T1E750 A0A182IJ93 A0A182YH39 A0A1B1W254 A0A0A1X7F2 A0A182RVW8 Q5EI69 A0A1S3D5P4 A0A2M3Z3M6 A0A195F9G5 A0A182N9Y9 D5L0M7 A0A182PU94 A0A2M3Z3F0 W5JIT2 A0A1W4V117
Pubmed
EMBL
AK343189
BAM73879.1
GQ915320
ACZ97414.1
GU731533
ADE05583.1
+ More
BABH01000585 NWSH01001018 PCG73090.1 KM016723 AID54875.1 KC789745 AGO62000.1 KX443459 ASO98034.1 KY212035 ARA91599.1 KQ459299 KPJ01870.1 PCG73083.1 AK289278 BAM73805.1 AK289277 BAM73804.1 AK343223 AK343224 BAM73905.1 PCG73091.1 DQ443362 ABF51451.1 KP001144 AJN91189.1 KM016725 AID54877.1 ODYU01001065 SOQ36787.1 RSAL01000074 RVE48932.1 BABH01000583 BABH01000584 KQ460761 KPJ12540.1 RVE48931.1 KF701160 AHW57330.1 JTDY01002470 KOB71350.1 AK289346 BAM73873.1 GFDF01000603 JAV13481.1 KF701161 AHW57331.1 CH477407 EAT41555.1 GFDL01004903 JAV30142.1 CH477503 EAT39885.1 GFDL01004915 JAV30130.1 GFDL01004861 JAV30184.1 GFDL01004871 JAV30174.1 JXUM01011183 JXUM01011184 KQ560325 KXJ83027.1 DS233130 EDS28283.1 NNAY01000453 OXU28285.1 KY997062 ATY49602.1 PCG73089.1 AAAB01008963 EAL39767.4 APCN01004898 AXCM01012454 AXCN02001347 KP004246 AJF45774.1 ATLV01017222 KE525157 KFB42070.1 AJWK01022137 AJWK01022138 AJWK01022139 AJWK01022140 AJWK01022141 AJWK01022142 AJWK01022143 ADMH02000163 ETN67528.1 GBYB01013159 JAG82926.1 AAAB01008885 EAL40625.3 EGK97044.1 EGK97045.1 APCN01008038 APCN01008039 CVRI01000019 CRK90412.1 KF656702 AHB59619.1 DQ117464 AAZ94273.1 DS232046 EDS33030.1 GFDL01005140 JAV29905.1 NEVH01002684 PNF41696.1 JRES01000290 KNC32693.1 JN984868 AFU75894.1 GFDL01005132 JAV29913.1 KP859400 AKZ17710.1 ODYU01002836 SOQ40811.1 KT601136 AME15805.1 CVRI01000036 CRK93146.1 AXCM01006317 GDHF01021566 JAI30748.1 KQ971338 EFA02880.1 ATLV01015123 KE525003 KFB40088.1 GAMD01003352 JAA98238.1 KU833274 ANW46746.1 GBXI01011493 GBXI01007225 JAD02799.1 JAD07067.1 AY880065 AAW78325.1 GGFM01002297 MBW23048.1 KQ981727 KYN37083.1 GU731532 ADE05582.1 GGFM01002300 MBW23051.1 ADMH02001125 ETN64031.1
BABH01000585 NWSH01001018 PCG73090.1 KM016723 AID54875.1 KC789745 AGO62000.1 KX443459 ASO98034.1 KY212035 ARA91599.1 KQ459299 KPJ01870.1 PCG73083.1 AK289278 BAM73805.1 AK289277 BAM73804.1 AK343223 AK343224 BAM73905.1 PCG73091.1 DQ443362 ABF51451.1 KP001144 AJN91189.1 KM016725 AID54877.1 ODYU01001065 SOQ36787.1 RSAL01000074 RVE48932.1 BABH01000583 BABH01000584 KQ460761 KPJ12540.1 RVE48931.1 KF701160 AHW57330.1 JTDY01002470 KOB71350.1 AK289346 BAM73873.1 GFDF01000603 JAV13481.1 KF701161 AHW57331.1 CH477407 EAT41555.1 GFDL01004903 JAV30142.1 CH477503 EAT39885.1 GFDL01004915 JAV30130.1 GFDL01004861 JAV30184.1 GFDL01004871 JAV30174.1 JXUM01011183 JXUM01011184 KQ560325 KXJ83027.1 DS233130 EDS28283.1 NNAY01000453 OXU28285.1 KY997062 ATY49602.1 PCG73089.1 AAAB01008963 EAL39767.4 APCN01004898 AXCM01012454 AXCN02001347 KP004246 AJF45774.1 ATLV01017222 KE525157 KFB42070.1 AJWK01022137 AJWK01022138 AJWK01022139 AJWK01022140 AJWK01022141 AJWK01022142 AJWK01022143 ADMH02000163 ETN67528.1 GBYB01013159 JAG82926.1 AAAB01008885 EAL40625.3 EGK97044.1 EGK97045.1 APCN01008038 APCN01008039 CVRI01000019 CRK90412.1 KF656702 AHB59619.1 DQ117464 AAZ94273.1 DS232046 EDS33030.1 GFDL01005140 JAV29905.1 NEVH01002684 PNF41696.1 JRES01000290 KNC32693.1 JN984868 AFU75894.1 GFDL01005132 JAV29913.1 KP859400 AKZ17710.1 ODYU01002836 SOQ40811.1 KT601136 AME15805.1 CVRI01000036 CRK93146.1 AXCM01006317 GDHF01021566 JAI30748.1 KQ971338 EFA02880.1 ATLV01015123 KE525003 KFB40088.1 GAMD01003352 JAA98238.1 KU833274 ANW46746.1 GBXI01011493 GBXI01007225 JAD02799.1 JAD07067.1 AY880065 AAW78325.1 GGFM01002297 MBW23048.1 KQ981727 KYN37083.1 GU731532 ADE05582.1 GGFM01002300 MBW23051.1 ADMH02001125 ETN64031.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000053240
UP000037510
+ More
UP000008820 UP000069940 UP000249989 UP000002320 UP000002358 UP000215335 UP000075900 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000076407 UP000075883 UP000069272 UP000075886 UP000075880 UP000075901 UP000030765 UP000075920 UP000076408 UP000075885 UP000075881 UP000092461 UP000075884 UP000000673 UP000183832 UP000235965 UP000037069 UP000007266 UP000079169 UP000078541 UP000192221
UP000008820 UP000069940 UP000249989 UP000002320 UP000002358 UP000215335 UP000075900 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000076407 UP000075883 UP000069272 UP000075886 UP000075880 UP000075901 UP000030765 UP000075920 UP000076408 UP000075885 UP000075881 UP000092461 UP000075884 UP000000673 UP000183832 UP000235965 UP000037069 UP000007266 UP000079169 UP000078541 UP000192221
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
L0N4R3
D2JLK4
D5L0M8
H9IUS3
A0A2A4JNU7
A0A068F0V7
+ More
S4VAQ8 A0A248QEH6 A0A1V0D9D0 A0A194QA50 A0A2A4JNR2 L0N788 L0N4H7 L0N7D2 A0A2A4JM80 Q1HPP0 A0A0C5C1N9 A0A068ETT4 A0A2H1V7J5 A0A3S2TKW7 H9IUS4 A0A194R5M7 A0A3S2P026 X5DB06 A0A0L7L7K6 L0N5J9 A0A1L8E432 X5D4H8 Q174P3 A0A1S4FEN8 A0A1Q3FRN6 Q16Z24 A0A1Q3FR81 A0A1Q3FRH5 A0A1Q3FRH0 A0A182GGE4 B0XHB7 K7J4F5 A0A232FC79 A0A2S0E492 A0A2A4JMP1 A0A182RYV2 A0A182UQZ2 A0A182U8B6 A0A182KW88 Q5TQQ1 A0A182HMS7 A0A182WY36 A0A182MQZ0 A0A182FJZ7 A0A182Q0J4 A0A182J321 A0A182TCR7 A0A0B5HSJ6 A0A084VVS6 A0A182W664 A0A182XYA1 A0A182P0M1 A0A182KG08 A0A1B0GJR2 A0A182MZX3 W5JVS3 A0A0C9R9S5 F5HL83 A0A182IDW2 A0A182VA75 A0A1J1HSQ0 V5T6Q9 Q3I413 B0WQV0 A0A1Q3FQS4 A0A2J7RLH7 A0A0L0CJW1 A0A1L1V8K0 A0A1Q3FR66 A0A0K1YWB8 A0A2H1VJ39 A0A125S6P4 A0A1J1I034 A0A182LRX0 A0A0K8UW61 A0A182W5Z8 D1ZZJ8 A0A084VQ44 T1E750 A0A182IJ93 A0A182YH39 A0A1B1W254 A0A0A1X7F2 A0A182RVW8 Q5EI69 A0A1S3D5P4 A0A2M3Z3M6 A0A195F9G5 A0A182N9Y9 D5L0M7 A0A182PU94 A0A2M3Z3F0 W5JIT2 A0A1W4V117
S4VAQ8 A0A248QEH6 A0A1V0D9D0 A0A194QA50 A0A2A4JNR2 L0N788 L0N4H7 L0N7D2 A0A2A4JM80 Q1HPP0 A0A0C5C1N9 A0A068ETT4 A0A2H1V7J5 A0A3S2TKW7 H9IUS4 A0A194R5M7 A0A3S2P026 X5DB06 A0A0L7L7K6 L0N5J9 A0A1L8E432 X5D4H8 Q174P3 A0A1S4FEN8 A0A1Q3FRN6 Q16Z24 A0A1Q3FR81 A0A1Q3FRH5 A0A1Q3FRH0 A0A182GGE4 B0XHB7 K7J4F5 A0A232FC79 A0A2S0E492 A0A2A4JMP1 A0A182RYV2 A0A182UQZ2 A0A182U8B6 A0A182KW88 Q5TQQ1 A0A182HMS7 A0A182WY36 A0A182MQZ0 A0A182FJZ7 A0A182Q0J4 A0A182J321 A0A182TCR7 A0A0B5HSJ6 A0A084VVS6 A0A182W664 A0A182XYA1 A0A182P0M1 A0A182KG08 A0A1B0GJR2 A0A182MZX3 W5JVS3 A0A0C9R9S5 F5HL83 A0A182IDW2 A0A182VA75 A0A1J1HSQ0 V5T6Q9 Q3I413 B0WQV0 A0A1Q3FQS4 A0A2J7RLH7 A0A0L0CJW1 A0A1L1V8K0 A0A1Q3FR66 A0A0K1YWB8 A0A2H1VJ39 A0A125S6P4 A0A1J1I034 A0A182LRX0 A0A0K8UW61 A0A182W5Z8 D1ZZJ8 A0A084VQ44 T1E750 A0A182IJ93 A0A182YH39 A0A1B1W254 A0A0A1X7F2 A0A182RVW8 Q5EI69 A0A1S3D5P4 A0A2M3Z3M6 A0A195F9G5 A0A182N9Y9 D5L0M7 A0A182PU94 A0A2M3Z3F0 W5JIT2 A0A1W4V117
PDB
6C94
E-value=3.1764e-58,
Score=571
Ontologies
GO
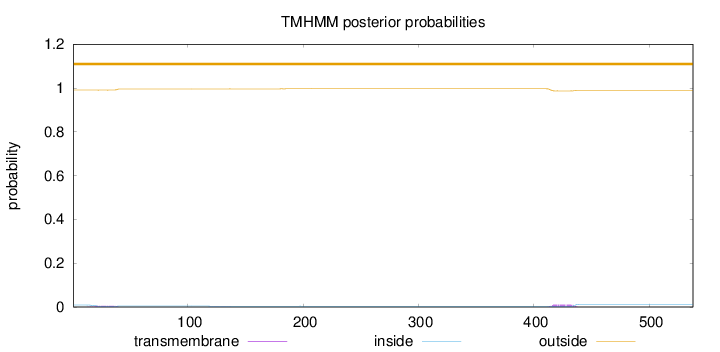
Topology
Length:
538
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.33936
Exp number, first 60 AAs:
0.10809
Total prob of N-in:
0.00912
outside
1 - 538
Population Genetic Test Statistics
Pi
205.646866
Theta
171.828195
Tajima's D
-1.002019
CLR
0.830932
CSRT
0.135793210339483
Interpretation
Uncertain
