Gene
KWMTBOMO07974
Pre Gene Modal
BGIBMGA001148
Annotation
achaete-scute-like_protein_ASH2_[Bombyx_mori]
Full name
Achaete-scute complex protein T3
Alternative Name
Protein lethal of scute
Transcription factor
Location in the cell
Nuclear Reliability : 4.235
Sequence
CDS
ATGCTCCAAGAAATCCAGCTAGTTCAAGGTCAGACGAATTACGTGGTCGTTTCTTCCGGATATCCTTCGAATACATTGAATAAATCATCTTTAGAGAAACGAAATGTTGCTATAGCTCCAGCGCCTGAGAAAAATTACGTCACACACGACACTCCACCGAATCTACACTACAGGAAAAAGGTGCATTTCAGAACTAATCCGTACACTGGACCACAGGCTGCGTCAATAGCGAGACGTAATGCACGGGAACGAAATCGCGTAAAACAAGTGAATGATGGATTCAACGCACTTCGCCGTCACCTGCCGGCTTCTGTCGTGGCAGCTCTGTCCGGTGGCGCCAGACGAGGTTCGTCAGGGAAGAAACTTAGCAAAGTCGACACATTACGGATGGTTGTTGAATATATAAGGTACCTTCAGCAGTTATTAGACGAAAGTGATGCCGCATTAGGTATTACGCGTGATCAAGAAAATCGGGAAAATATTCCAAGCAATAACTCAGTTCAGCCGATGACTTCTATTGACATGGATGACGGGTTTTTCTACGGAAGTGGATCACCTTGTTCAGAGAAGGCAGATTCGCCAGCTCCTTCGGAATGTTCTTCGGGTGTGTCTTCGGCGTATTCGGCTGTCGATCGTTACGAGGTTACTACGCAGCAACAAATGGGATCAATGGATGAAGAAGAACTCTTAGACGTCATTTCATGGTGGCAACAAAAATAG
Protein
MLQEIQLVQGQTNYVVVSSGYPSNTLNKSSLEKRNVAIAPAPEKNYVTHDTPPNLHYRKKVHFRTNPYTGPQAASIARRNARERNRVKQVNDGFNALRRHLPASVVAALSGGARRGSSGKKLSKVDTLRMVVEYIRYLQQLLDESDAALGITRDQENRENIPSNNSVQPMTSIDMDDGFFYGSGSPCSEKADSPAPSECSSGVSSAYSAVDRYEVTTQQQMGSMDEEELLDVISWWQQK
Summary
Subunit
Efficient DNA binding requires dimerization with another bHLH protein.
Miscellaneous
AS-C proteins are expressed first earlier in ectoderm and then in the internalized neuroblasts, while T8 is expressed in presumptive neural precursors, once they have segregated from the ectoderm.
Keywords
Complete proteome
Developmental protein
Differentiation
DNA-binding
Neurogenesis
Reference proteome
Feature
chain Achaete-scute complex protein T3
Uniprot
A6N867
A0A2A4J938
A0A2H1VLD1
O76488
A0A0L7KWQ5
A0A194Q8Y4
+ More
A0A212ENF6 A0A194R9L4 A0A0H3UYT4 A0A2H1VME4 A0A2A4J920 A0A0M4F949 A0A194QA55 A0A194R4E4 H9IUS0 A6N868 A0A212ENG6 Q16Z21 A0A182GS63 A0A0P6FRG6 A0A2P8YB04 A0A194Q8M1 A0A336KPN9 U4UNX8 A0A2A4J581 Q2I1Z2 H9IUS1 A0A2R7W5Z6 A0A212ENE2 A0A0N0BD42 A0A154PKB8 A0A1S4FJ51 Q16Z23 A0A310SMU7 A0A0H3UYS2 Q174P2 A0A0M4F923 A0A0L7KP59 N6TZG9 A0A0L7R3G1 A0A067R5E0 T1HC65 A3E0V8 E9IF61 A0A226EPU9 A0A1S3CTP3 Q6VYM9 Q8WQQ6 A0A0K8T2Y0 A0A1W4XHC9 A0A194R5N2 E0VXK7 B3MSE1 P09774 A0A0F7IKF2 A0A195CD83 O77032 Q9GNL5 Q9U910
A0A212ENF6 A0A194R9L4 A0A0H3UYT4 A0A2H1VME4 A0A2A4J920 A0A0M4F949 A0A194QA55 A0A194R4E4 H9IUS0 A6N868 A0A212ENG6 Q16Z21 A0A182GS63 A0A0P6FRG6 A0A2P8YB04 A0A194Q8M1 A0A336KPN9 U4UNX8 A0A2A4J581 Q2I1Z2 H9IUS1 A0A2R7W5Z6 A0A212ENE2 A0A0N0BD42 A0A154PKB8 A0A1S4FJ51 Q16Z23 A0A310SMU7 A0A0H3UYS2 Q174P2 A0A0M4F923 A0A0L7KP59 N6TZG9 A0A0L7R3G1 A0A067R5E0 T1HC65 A3E0V8 E9IF61 A0A226EPU9 A0A1S3CTP3 Q6VYM9 Q8WQQ6 A0A0K8T2Y0 A0A1W4XHC9 A0A194R5N2 E0VXK7 B3MSE1 P09774 A0A0F7IKF2 A0A195CD83 O77032 Q9GNL5 Q9U910
Pubmed
EMBL
BABH01000604
EF620927
ABR20839.1
NWSH01002585
PCG67913.1
ODYU01003190
+ More
SOQ41611.1 AF071498 AAC24714.1 JTDY01004860 KOB67667.1 KQ459299 KPJ01874.1 AGBW02013662 OWR43024.1 KQ460761 KPJ12546.1 KJ158243 AIZ67913.1 SOQ41612.1 PCG67912.1 KP245732 ALC76152.1 KPJ01875.1 KPJ12547.1 BABH01000608 EF620928 ABR20840.1 OWR43023.1 CH477503 EAT39888.1 JXUM01085146 KQ563483 KXJ73762.1 GDIQ01045359 JAN49378.1 PYGN01000741 PSN41442.1 KPJ01873.1 UFQS01000677 UFQT01000677 SSX06121.1 SSX26477.1 KB632299 ERL91735.1 NWSH01003306 PCG66590.1 DQ350889 DQ350890 ABC84346.1 ABC84347.1 BABH01000596 KK854372 PTY15157.1 OWR43025.1 KQ435878 KOX69918.1 KQ434946 KZC12321.1 EAT39886.1 KQ761046 OAD58328.1 KJ158244 AIZ67914.1 CH477407 EAT41556.1 CP012528 ALC48632.1 JTDY01008101 KOB64739.1 APGK01002009 KB734570 ENN83543.1 KQ414663 KOC65425.1 KK852878 KDR14510.1 ACPB03007141 DQ489559 ABF50840.2 GL762758 EFZ20812.1 LNIX01000002 OXA59177.1 AY319382 AAQ82735.1 AY061876 AAL32066.1 GBRD01005924 GDHC01005821 JAG59897.1 JAQ12808.1 KPJ12545.1 DS235833 EEB18113.1 CH902622 EDV34696.2 X12549 X71806 AE014298 AL024453 AY121693 KP147933 AKG92770.1 KQ978009 KYM98171.1 AB005802 CM000366 BAA33213.1 EDX16748.1 AB032461 BAB20411.1 AB017570 CH954183 BAA86616.1 EDV45356.1
SOQ41611.1 AF071498 AAC24714.1 JTDY01004860 KOB67667.1 KQ459299 KPJ01874.1 AGBW02013662 OWR43024.1 KQ460761 KPJ12546.1 KJ158243 AIZ67913.1 SOQ41612.1 PCG67912.1 KP245732 ALC76152.1 KPJ01875.1 KPJ12547.1 BABH01000608 EF620928 ABR20840.1 OWR43023.1 CH477503 EAT39888.1 JXUM01085146 KQ563483 KXJ73762.1 GDIQ01045359 JAN49378.1 PYGN01000741 PSN41442.1 KPJ01873.1 UFQS01000677 UFQT01000677 SSX06121.1 SSX26477.1 KB632299 ERL91735.1 NWSH01003306 PCG66590.1 DQ350889 DQ350890 ABC84346.1 ABC84347.1 BABH01000596 KK854372 PTY15157.1 OWR43025.1 KQ435878 KOX69918.1 KQ434946 KZC12321.1 EAT39886.1 KQ761046 OAD58328.1 KJ158244 AIZ67914.1 CH477407 EAT41556.1 CP012528 ALC48632.1 JTDY01008101 KOB64739.1 APGK01002009 KB734570 ENN83543.1 KQ414663 KOC65425.1 KK852878 KDR14510.1 ACPB03007141 DQ489559 ABF50840.2 GL762758 EFZ20812.1 LNIX01000002 OXA59177.1 AY319382 AAQ82735.1 AY061876 AAL32066.1 GBRD01005924 GDHC01005821 JAG59897.1 JAQ12808.1 KPJ12545.1 DS235833 EEB18113.1 CH902622 EDV34696.2 X12549 X71806 AE014298 AL024453 AY121693 KP147933 AKG92770.1 KQ978009 KYM98171.1 AB005802 CM000366 BAA33213.1 EDX16748.1 AB032461 BAB20411.1 AB017570 CH954183 BAA86616.1 EDV45356.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000007151
UP000053240
+ More
UP000008820 UP000069940 UP000249989 UP000245037 UP000030742 UP000053105 UP000076502 UP000092553 UP000019118 UP000053825 UP000027135 UP000015103 UP000198287 UP000079169 UP000192223 UP000009046 UP000007801 UP000000803 UP000078542 UP000000304 UP000008711
UP000008820 UP000069940 UP000249989 UP000245037 UP000030742 UP000053105 UP000076502 UP000092553 UP000019118 UP000053825 UP000027135 UP000015103 UP000198287 UP000079169 UP000192223 UP000009046 UP000007801 UP000000803 UP000078542 UP000000304 UP000008711
Pfam
PF00010 HLH
SUPFAM
SSF47459
SSF47459
Gene 3D
CDD
ProteinModelPortal
A6N867
A0A2A4J938
A0A2H1VLD1
O76488
A0A0L7KWQ5
A0A194Q8Y4
+ More
A0A212ENF6 A0A194R9L4 A0A0H3UYT4 A0A2H1VME4 A0A2A4J920 A0A0M4F949 A0A194QA55 A0A194R4E4 H9IUS0 A6N868 A0A212ENG6 Q16Z21 A0A182GS63 A0A0P6FRG6 A0A2P8YB04 A0A194Q8M1 A0A336KPN9 U4UNX8 A0A2A4J581 Q2I1Z2 H9IUS1 A0A2R7W5Z6 A0A212ENE2 A0A0N0BD42 A0A154PKB8 A0A1S4FJ51 Q16Z23 A0A310SMU7 A0A0H3UYS2 Q174P2 A0A0M4F923 A0A0L7KP59 N6TZG9 A0A0L7R3G1 A0A067R5E0 T1HC65 A3E0V8 E9IF61 A0A226EPU9 A0A1S3CTP3 Q6VYM9 Q8WQQ6 A0A0K8T2Y0 A0A1W4XHC9 A0A194R5N2 E0VXK7 B3MSE1 P09774 A0A0F7IKF2 A0A195CD83 O77032 Q9GNL5 Q9U910
A0A212ENF6 A0A194R9L4 A0A0H3UYT4 A0A2H1VME4 A0A2A4J920 A0A0M4F949 A0A194QA55 A0A194R4E4 H9IUS0 A6N868 A0A212ENG6 Q16Z21 A0A182GS63 A0A0P6FRG6 A0A2P8YB04 A0A194Q8M1 A0A336KPN9 U4UNX8 A0A2A4J581 Q2I1Z2 H9IUS1 A0A2R7W5Z6 A0A212ENE2 A0A0N0BD42 A0A154PKB8 A0A1S4FJ51 Q16Z23 A0A310SMU7 A0A0H3UYS2 Q174P2 A0A0M4F923 A0A0L7KP59 N6TZG9 A0A0L7R3G1 A0A067R5E0 T1HC65 A3E0V8 E9IF61 A0A226EPU9 A0A1S3CTP3 Q6VYM9 Q8WQQ6 A0A0K8T2Y0 A0A1W4XHC9 A0A194R5N2 E0VXK7 B3MSE1 P09774 A0A0F7IKF2 A0A195CD83 O77032 Q9GNL5 Q9U910
Ontologies
KEGG
GO
PANTHER
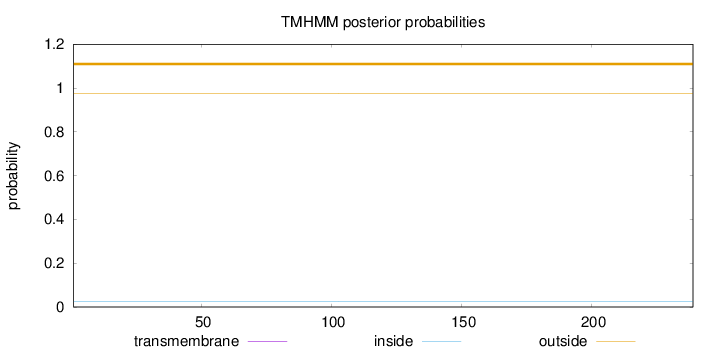
Topology
Length:
239
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00272
Exp number, first 60 AAs:
0.00102
Total prob of N-in:
0.02601
outside
1 - 239
Population Genetic Test Statistics
Pi
157.49746
Theta
177.033801
Tajima's D
-0.30401
CLR
1.001496
CSRT
0.287035648217589
Interpretation
Uncertain
