Gene
KWMTBOMO07970
Pre Gene Modal
BGIBMGA001151
Annotation
PREDICTED:_proton-coupled_amino_acid_transporter_1-like_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 4.901
Sequence
CDS
ATGAGTGGGAAGACTGTTAAGGAAGACGAATACAATCCGTATGAACATCGTAAAATCGAGAAGCCAAATTCAGATATCAGGTCATTAGCGAATCTGATTAAATCTTCGTTGGGCTCTGGCCTACTTGCTATGCCACTGGCTTTCTCCAATTCGGGCTGGGCAGTCGGCATTATTGGAACAATCGTCATTGCATTTGTTTGTGGACACTGTGTGCATATATTCGTGAAAACATCAAGAGGATGTTGCAAGCTCGTGCGACGACCTCTTCTAAGCTACTCGGAGACTTGCAAAGCTGCATTCGAGCATGGACCAAAGTCGTTACGACCTTTCGCTTTTGCTGCACAATTATTTTCCGAATTTGCCTTATTCTTCACCTACATCGGTGTTTGCTGTATTTTTACCGTTCTAATTGCTGATTCTTTAAAACAGTTGTTCGATGAATACGTCACTACAACAGTCCTGCCTGTGGAATATTATTGTTTAATCATACTCGTGCCACTTTGCCTGATGGTACAGATCAGGCACTTAAAATGGTTGGCACCGTTTTCATTTATAGCAAACATTCTACTGATTGCAACTTTCGTTATCTGCATGTACTATATATTTGGAGACGAAATTAATTTTACTGATAAAAGAATCTCAGGGGATCTGTCTAGACTCCCGGCTTTTTTGTCCACGGTTATATTTGCGATGGAAGGCATCGGAGTAGTGATGCCAGTTGAAAATACAATGAAAAAACCGCAGCATTTTCTAGGGTGCCCTAGTGTCCTCGTCGTAGCTATGTCAACTATCATGCTCATGTATTCTATTCTTGGCTTATTCGGCTACTTCAGATACGGAGATGTACTTAGAGGATCTATAACACTGAACCTACCTATTGACGACTGGCCAGCCGTGTGCGCTAAAACTTTTATAGCACTATCAATATTCTTTACATACCCACTTCACTTCTTTGTGGTACTAGATATATTCACCCGATATGCGGAACCACACATCAAGAAGGAGTATCGTAATTTTGCACAAATACTCGCCAGGACCTCTTGCGTTTGGATTTGTGGTGGAATAGGCATAGCATTGCCTATGCTGGAACAGATCATCAACATTGTCGGAGCACTGTTCTACTCCATACTGGGACTCATAATACCAGGTGTAATCGAGACAGTATTTAGATGGGAAGATCTGGGCAGATGGAATTGGATTTTTTGGAAAAATCTATTGATTGTCCTCTTTGGTGTCTGTTCTTTAGTATCAGGCTGTACTGTTAGTGTCATGGATATTATAAGTATTTTAAATAAAAAAACCATTTGA
Protein
MSGKTVKEDEYNPYEHRKIEKPNSDIRSLANLIKSSLGSGLLAMPLAFSNSGWAVGIIGTIVIAFVCGHCVHIFVKTSRGCCKLVRRPLLSYSETCKAAFEHGPKSLRPFAFAAQLFSEFALFFTYIGVCCIFTVLIADSLKQLFDEYVTTTVLPVEYYCLIILVPLCLMVQIRHLKWLAPFSFIANILLIATFVICMYYIFGDEINFTDKRISGDLSRLPAFLSTVIFAMEGIGVVMPVENTMKKPQHFLGCPSVLVVAMSTIMLMYSILGLFGYFRYGDVLRGSITLNLPIDDWPAVCAKTFIALSIFFTYPLHFFVVLDIFTRYAEPHIKKEYRNFAQILARTSCVWICGGIGIALPMLEQIINIVGALFYSILGLIIPGVIETVFRWEDLGRWNWIFWKNLLIVLFGVCSLVSGCTVSVMDIISILNKKTI
Summary
Cofactor
FAD
Uniprot
A0A2H1V1S3
A0A194R4V9
S4PGT3
A0A2A4JI41
A0A194Q8M5
A0A2A4JJQ0
+ More
A0A194Q8L4 A0A212ENJ6 A0A212ENE9 A0A2H1VMY1 A0A212FJG4 A0A194PCR8 I4DJF9 A0A182Q0A8 A0A212EWW9 A0A2A4JJ72 A0A182IP57 W5JSE3 A0A182USW0 A0A1S4H4R1 A0A182KV18 A0A182MBQ3 A0A1E1W5A1 T1EA00 A0A2M4A0X7 A0A2M3YY91 A0A2M4A0Y6 A0A2M4BME5 A0A182YAD2 A0A2M4A1D4 B0WV85 A0A336MRI7 H9IUB4 A0A1Q3F2I9 A0A1L8DEN1 A0A1B0CAN0 Q17E30 A0A194PJ35 A0A194Q8Y9 A0A194QKK0 A0A2A4JEW1 A0A212F4Y3 A0A182UK13 A0A0M9A0S8 A0A182RLT5 A0A1L8DEV4 A0A182K1E2 A0A182WXY0 W8BC78 A0A182I602 A0A182WE65 A0A182FPM2 T1PE56 A0A1Y1LDI1 A0A1I8PFI4 A0A0L7QKA8 A0A182NPB8 A0A1E1WJX2 B4KSQ0 A0A1E1WI99 A0A1L8EGD4 A0A1E1W9Z5 A0A0L0BM39 D3TMU6 W8BH26 A0A1B0A1Z2 A0A0T6AW13 A0A0K8UWL5 B3NRX3 B4MPP1 A0A1W4VPX7 A0A034VK49 B4P566 H9IVL9 A0A2J7PFJ4 I4DNP1 B3MCU4 A0A0Q5W221 A0A0R1DWD8 A0A3B0JQL4 A0A1B0BDW6 Q7K2W3 A0A0M4EAI4 B4QDE8 B4GBL8 Q293E9 B4HPM4 A0A194R9L9 A0A1J1HGM7 A0A212FDX9 A0A194PEI1 B4LP82 A0A067RHJ6 A0A1Y1LSA3 A0A3S2N8W7 A0A1Y1K9F3 A0A1Y1K9N8 B4JVM3
A0A194Q8L4 A0A212ENJ6 A0A212ENE9 A0A2H1VMY1 A0A212FJG4 A0A194PCR8 I4DJF9 A0A182Q0A8 A0A212EWW9 A0A2A4JJ72 A0A182IP57 W5JSE3 A0A182USW0 A0A1S4H4R1 A0A182KV18 A0A182MBQ3 A0A1E1W5A1 T1EA00 A0A2M4A0X7 A0A2M3YY91 A0A2M4A0Y6 A0A2M4BME5 A0A182YAD2 A0A2M4A1D4 B0WV85 A0A336MRI7 H9IUB4 A0A1Q3F2I9 A0A1L8DEN1 A0A1B0CAN0 Q17E30 A0A194PJ35 A0A194Q8Y9 A0A194QKK0 A0A2A4JEW1 A0A212F4Y3 A0A182UK13 A0A0M9A0S8 A0A182RLT5 A0A1L8DEV4 A0A182K1E2 A0A182WXY0 W8BC78 A0A182I602 A0A182WE65 A0A182FPM2 T1PE56 A0A1Y1LDI1 A0A1I8PFI4 A0A0L7QKA8 A0A182NPB8 A0A1E1WJX2 B4KSQ0 A0A1E1WI99 A0A1L8EGD4 A0A1E1W9Z5 A0A0L0BM39 D3TMU6 W8BH26 A0A1B0A1Z2 A0A0T6AW13 A0A0K8UWL5 B3NRX3 B4MPP1 A0A1W4VPX7 A0A034VK49 B4P566 H9IVL9 A0A2J7PFJ4 I4DNP1 B3MCU4 A0A0Q5W221 A0A0R1DWD8 A0A3B0JQL4 A0A1B0BDW6 Q7K2W3 A0A0M4EAI4 B4QDE8 B4GBL8 Q293E9 B4HPM4 A0A194R9L9 A0A1J1HGM7 A0A212FDX9 A0A194PEI1 B4LP82 A0A067RHJ6 A0A1Y1LSA3 A0A3S2N8W7 A0A1Y1K9F3 A0A1Y1K9N8 B4JVM3
Pubmed
26354079
23622113
22118469
22651552
20920257
23761445
+ More
20966253 25244985 19121390 17510324 24495485 25315136 28004739 17994087 18057021 26108605 20353571 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 23185243 24845553
20966253 25244985 19121390 17510324 24495485 25315136 28004739 17994087 18057021 26108605 20353571 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 23185243 24845553
EMBL
ODYU01000291
SOQ34787.1
KQ460761
KPJ12549.1
GAIX01003512
JAA89048.1
+ More
NWSH01001335 PCG71631.1 KQ459299 KPJ01878.1 PCG71630.1 KPJ01877.1 AGBW02013662 OWR43021.1 OWR43020.1 ODYU01003190 SOQ41614.1 AGBW02008267 OWR53876.1 KQ459606 KPI91091.1 AK401427 BAM18049.1 AXCN02001819 AGBW02011868 OWR45996.1 PCG71634.1 ADMH02000303 ETN67041.1 AXCM01002773 GDQN01008899 JAT82155.1 GAMD01000852 JAB00739.1 GGFK01001081 MBW34402.1 GGFM01000469 MBW21220.1 GGFK01001091 MBW34412.1 GGFJ01005061 MBW54202.1 GGFK01001241 MBW34562.1 DS232119 EDS35424.1 UFQT01001035 SSX28568.1 BABH01001422 BABH01001423 GFDL01013271 JAV21774.1 GFDF01009178 JAV04906.1 AJWK01004116 AJWK01004117 AJWK01004118 AJWK01004119 CH477287 EAT44708.1 KPI91095.1 KPJ01879.1 KQ461198 KPJ06088.1 NWSH01001829 PCG69980.1 AGBW02010286 OWR48805.1 KQ435775 KOX75035.1 GFDF01009177 JAV04907.1 GAMC01010263 JAB96292.1 APCN01003539 KA647011 AFP61640.1 GEZM01061537 JAV70400.1 KQ414957 KOC59052.1 GDQN01003847 JAT87207.1 CH933808 EDW10549.1 KRG05311.1 GDQN01004339 JAT86715.1 GFDG01001017 JAV17782.1 GDQN01007289 JAT83765.1 JRES01001654 KNC21116.1 EZ422748 ADD19024.1 GAMC01010262 GAMC01010261 JAB96293.1 LJIG01022698 KRT79188.1 GDHF01021391 JAI30923.1 CH954179 EDV56275.1 KQS62807.1 CH963849 EDW74080.1 GAKP01016792 GAKP01016790 JAC42160.1 CM000158 EDW90728.1 KRJ99410.1 BABH01001445 BABH01001446 BABH01001447 NEVH01025657 PNF15087.1 AK403058 BAM19531.1 CH902619 EDV37346.2 KPU76737.1 KQS62808.1 KRJ99411.1 OUUW01000001 SPP75939.1 JXJN01012713 AE013599 AY060621 AAF58478.2 AAL28169.1 CP012524 ALC42096.1 CM000362 CM002911 EDX06813.1 KMY93293.1 CH479181 EDW31313.1 CM000071 EAL24662.1 KRT01443.1 CH480816 EDW47608.1 KPJ12551.1 CVRI01000004 CRK87157.1 AGBW02008991 OWR51959.1 KPI91099.1 CH940648 EDW60191.2 KRF79294.1 KK852631 KDR19825.1 GEZM01052528 JAV74456.1 RSAL01000191 RVE44648.1 GEZM01088603 JAV58073.1 GEZM01088602 JAV58074.1 CH916375 EDV98491.1
NWSH01001335 PCG71631.1 KQ459299 KPJ01878.1 PCG71630.1 KPJ01877.1 AGBW02013662 OWR43021.1 OWR43020.1 ODYU01003190 SOQ41614.1 AGBW02008267 OWR53876.1 KQ459606 KPI91091.1 AK401427 BAM18049.1 AXCN02001819 AGBW02011868 OWR45996.1 PCG71634.1 ADMH02000303 ETN67041.1 AXCM01002773 GDQN01008899 JAT82155.1 GAMD01000852 JAB00739.1 GGFK01001081 MBW34402.1 GGFM01000469 MBW21220.1 GGFK01001091 MBW34412.1 GGFJ01005061 MBW54202.1 GGFK01001241 MBW34562.1 DS232119 EDS35424.1 UFQT01001035 SSX28568.1 BABH01001422 BABH01001423 GFDL01013271 JAV21774.1 GFDF01009178 JAV04906.1 AJWK01004116 AJWK01004117 AJWK01004118 AJWK01004119 CH477287 EAT44708.1 KPI91095.1 KPJ01879.1 KQ461198 KPJ06088.1 NWSH01001829 PCG69980.1 AGBW02010286 OWR48805.1 KQ435775 KOX75035.1 GFDF01009177 JAV04907.1 GAMC01010263 JAB96292.1 APCN01003539 KA647011 AFP61640.1 GEZM01061537 JAV70400.1 KQ414957 KOC59052.1 GDQN01003847 JAT87207.1 CH933808 EDW10549.1 KRG05311.1 GDQN01004339 JAT86715.1 GFDG01001017 JAV17782.1 GDQN01007289 JAT83765.1 JRES01001654 KNC21116.1 EZ422748 ADD19024.1 GAMC01010262 GAMC01010261 JAB96293.1 LJIG01022698 KRT79188.1 GDHF01021391 JAI30923.1 CH954179 EDV56275.1 KQS62807.1 CH963849 EDW74080.1 GAKP01016792 GAKP01016790 JAC42160.1 CM000158 EDW90728.1 KRJ99410.1 BABH01001445 BABH01001446 BABH01001447 NEVH01025657 PNF15087.1 AK403058 BAM19531.1 CH902619 EDV37346.2 KPU76737.1 KQS62808.1 KRJ99411.1 OUUW01000001 SPP75939.1 JXJN01012713 AE013599 AY060621 AAF58478.2 AAL28169.1 CP012524 ALC42096.1 CM000362 CM002911 EDX06813.1 KMY93293.1 CH479181 EDW31313.1 CM000071 EAL24662.1 KRT01443.1 CH480816 EDW47608.1 KPJ12551.1 CVRI01000004 CRK87157.1 AGBW02008991 OWR51959.1 KPI91099.1 CH940648 EDW60191.2 KRF79294.1 KK852631 KDR19825.1 GEZM01052528 JAV74456.1 RSAL01000191 RVE44648.1 GEZM01088603 JAV58073.1 GEZM01088602 JAV58074.1 CH916375 EDV98491.1
Proteomes
UP000053240
UP000218220
UP000053268
UP000007151
UP000075886
UP000075880
+ More
UP000000673 UP000075903 UP000075882 UP000075883 UP000076408 UP000002320 UP000005204 UP000092461 UP000008820 UP000075902 UP000053105 UP000075900 UP000075881 UP000076407 UP000075840 UP000075920 UP000069272 UP000095301 UP000095300 UP000053825 UP000075884 UP000009192 UP000037069 UP000092445 UP000008711 UP000007798 UP000192221 UP000002282 UP000235965 UP000007801 UP000268350 UP000092460 UP000000803 UP000092553 UP000000304 UP000008744 UP000001819 UP000001292 UP000183832 UP000008792 UP000027135 UP000283053 UP000001070
UP000000673 UP000075903 UP000075882 UP000075883 UP000076408 UP000002320 UP000005204 UP000092461 UP000008820 UP000075902 UP000053105 UP000075900 UP000075881 UP000076407 UP000075840 UP000075920 UP000069272 UP000095301 UP000095300 UP000053825 UP000075884 UP000009192 UP000037069 UP000092445 UP000008711 UP000007798 UP000192221 UP000002282 UP000235965 UP000007801 UP000268350 UP000092460 UP000000803 UP000092553 UP000000304 UP000008744 UP000001819 UP000001292 UP000183832 UP000008792 UP000027135 UP000283053 UP000001070
Interpro
IPR013057
AA_transpt_TM
+ More
IPR029846 Yellow
IPR011042 6-blade_b-propeller_TolB-like
IPR017996 Royal_jelly/protein_yellow
IPR001834 CBR-like
IPR017938 Riboflavin_synthase-like_b-brl
IPR001433 OxRdtase_FAD/NAD-bd
IPR017927 FAD-bd_FR_type
IPR008333 Cbr1-like_FAD-bd_dom
IPR039261 FNR_nucleotide-bd
IPR001709 Flavoprot_Pyr_Nucl_cyt_Rdtase
IPR029846 Yellow
IPR011042 6-blade_b-propeller_TolB-like
IPR017996 Royal_jelly/protein_yellow
IPR001834 CBR-like
IPR017938 Riboflavin_synthase-like_b-brl
IPR001433 OxRdtase_FAD/NAD-bd
IPR017927 FAD-bd_FR_type
IPR008333 Cbr1-like_FAD-bd_dom
IPR039261 FNR_nucleotide-bd
IPR001709 Flavoprot_Pyr_Nucl_cyt_Rdtase
Gene 3D
ProteinModelPortal
A0A2H1V1S3
A0A194R4V9
S4PGT3
A0A2A4JI41
A0A194Q8M5
A0A2A4JJQ0
+ More
A0A194Q8L4 A0A212ENJ6 A0A212ENE9 A0A2H1VMY1 A0A212FJG4 A0A194PCR8 I4DJF9 A0A182Q0A8 A0A212EWW9 A0A2A4JJ72 A0A182IP57 W5JSE3 A0A182USW0 A0A1S4H4R1 A0A182KV18 A0A182MBQ3 A0A1E1W5A1 T1EA00 A0A2M4A0X7 A0A2M3YY91 A0A2M4A0Y6 A0A2M4BME5 A0A182YAD2 A0A2M4A1D4 B0WV85 A0A336MRI7 H9IUB4 A0A1Q3F2I9 A0A1L8DEN1 A0A1B0CAN0 Q17E30 A0A194PJ35 A0A194Q8Y9 A0A194QKK0 A0A2A4JEW1 A0A212F4Y3 A0A182UK13 A0A0M9A0S8 A0A182RLT5 A0A1L8DEV4 A0A182K1E2 A0A182WXY0 W8BC78 A0A182I602 A0A182WE65 A0A182FPM2 T1PE56 A0A1Y1LDI1 A0A1I8PFI4 A0A0L7QKA8 A0A182NPB8 A0A1E1WJX2 B4KSQ0 A0A1E1WI99 A0A1L8EGD4 A0A1E1W9Z5 A0A0L0BM39 D3TMU6 W8BH26 A0A1B0A1Z2 A0A0T6AW13 A0A0K8UWL5 B3NRX3 B4MPP1 A0A1W4VPX7 A0A034VK49 B4P566 H9IVL9 A0A2J7PFJ4 I4DNP1 B3MCU4 A0A0Q5W221 A0A0R1DWD8 A0A3B0JQL4 A0A1B0BDW6 Q7K2W3 A0A0M4EAI4 B4QDE8 B4GBL8 Q293E9 B4HPM4 A0A194R9L9 A0A1J1HGM7 A0A212FDX9 A0A194PEI1 B4LP82 A0A067RHJ6 A0A1Y1LSA3 A0A3S2N8W7 A0A1Y1K9F3 A0A1Y1K9N8 B4JVM3
A0A194Q8L4 A0A212ENJ6 A0A212ENE9 A0A2H1VMY1 A0A212FJG4 A0A194PCR8 I4DJF9 A0A182Q0A8 A0A212EWW9 A0A2A4JJ72 A0A182IP57 W5JSE3 A0A182USW0 A0A1S4H4R1 A0A182KV18 A0A182MBQ3 A0A1E1W5A1 T1EA00 A0A2M4A0X7 A0A2M3YY91 A0A2M4A0Y6 A0A2M4BME5 A0A182YAD2 A0A2M4A1D4 B0WV85 A0A336MRI7 H9IUB4 A0A1Q3F2I9 A0A1L8DEN1 A0A1B0CAN0 Q17E30 A0A194PJ35 A0A194Q8Y9 A0A194QKK0 A0A2A4JEW1 A0A212F4Y3 A0A182UK13 A0A0M9A0S8 A0A182RLT5 A0A1L8DEV4 A0A182K1E2 A0A182WXY0 W8BC78 A0A182I602 A0A182WE65 A0A182FPM2 T1PE56 A0A1Y1LDI1 A0A1I8PFI4 A0A0L7QKA8 A0A182NPB8 A0A1E1WJX2 B4KSQ0 A0A1E1WI99 A0A1L8EGD4 A0A1E1W9Z5 A0A0L0BM39 D3TMU6 W8BH26 A0A1B0A1Z2 A0A0T6AW13 A0A0K8UWL5 B3NRX3 B4MPP1 A0A1W4VPX7 A0A034VK49 B4P566 H9IVL9 A0A2J7PFJ4 I4DNP1 B3MCU4 A0A0Q5W221 A0A0R1DWD8 A0A3B0JQL4 A0A1B0BDW6 Q7K2W3 A0A0M4EAI4 B4QDE8 B4GBL8 Q293E9 B4HPM4 A0A194R9L9 A0A1J1HGM7 A0A212FDX9 A0A194PEI1 B4LP82 A0A067RHJ6 A0A1Y1LSA3 A0A3S2N8W7 A0A1Y1K9F3 A0A1Y1K9N8 B4JVM3
Ontologies
PANTHER
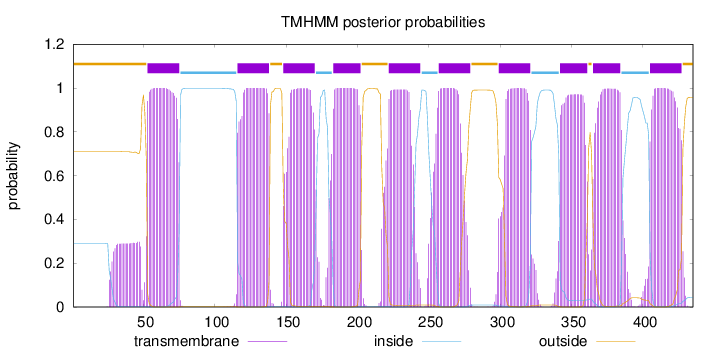
Topology
Length:
435
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
224.3489
Exp number, first 60 AAs:
14.68599
Total prob of N-in:
0.29058
POSSIBLE N-term signal
sequence
outside
1 - 52
TMhelix
53 - 75
inside
76 - 115
TMhelix
116 - 138
outside
139 - 147
TMhelix
148 - 170
inside
171 - 182
TMhelix
183 - 202
outside
203 - 221
TMhelix
222 - 244
inside
245 - 256
TMhelix
257 - 279
outside
280 - 298
TMhelix
299 - 321
inside
322 - 341
TMhelix
342 - 361
outside
362 - 364
TMhelix
365 - 384
inside
385 - 404
TMhelix
405 - 427
outside
428 - 435
Population Genetic Test Statistics
Pi
229.040852
Theta
185.930798
Tajima's D
0.54073
CLR
0.130741
CSRT
0.526923653817309
Interpretation
Uncertain
