Gene
KWMTBOMO07969 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001152
Annotation
PREDICTED:_NADH-cytochrome_b5_reductase_2_isoform_X1_[Bombyx_mori]
Full name
NADH-cytochrome b5 reductase
Location in the cell
Cytoplasmic Reliability : 1.167 Mitochondrial Reliability : 1.315 Nuclear Reliability : 1.127
Sequence
CDS
ATGTTTCAGACTATCAATTCTAAAATTGTACTACCAATAATATTTGGTGTTGGATCAGTGGTAGTCTTAGCAACTATAATAGCCAATTGTCTATGGGGAAAGAAATCTAAAGGATCTAGCAAAAAGTTAATAACATTAGTAGATTCTAACGTAAAATATGCATTACCGCTGATAGAAAGGGAAGAAATCAGTCATGACACCAGGAGATTCAGGCTGGGACTGCCGTCTCCTCAACATGTATTGGGATTACCGATCGGTCAACATATTCATTTGTCTGCAAAAATTAACGACGACCTAGTCATAAGAGCATACACGCCGGTGTCGAGCGATGAGGATAAAGGCTATGTCGATTTGGTTATCAAGGTATACTTTAAGAATGTACATCCGAAGTTTCCCGAGGGAGGGAAGCTGTCTCAGTACTTGAACAATATGAAGATAAATGATACTATAGATGTCCGTGGACCGTCAGGGAGATTGCAGTACACAGGAAACGGAACATTCCTCATTAAGAAATTAAGAAAAGATCCGCCTACAAAAGTTGTCGTCAAGAAATTGAACTTGATTGCAGGCGGTACTGGCATAGCTCCAATGCTTCAGCTGGTCCGACATATATGCACTGACGTCAACGACCGCACCGAACTGAAGCTGCTATTCGCGAACCAAAGCGAAGATGACATTCTACTTAGAGATGAATTAGAAAGATACCAAAGGGAACATCCTTCCCAATTCCAAGTTTGGTACACGATTGATCGGCCAACGGATGGTTGGAAATACAGCTCCGGTTTCATCAACGATGAAATGATCCGCGACCATTTGTTTCCGCCGTCCAACGATGTTCTAGTACTGATGTGCGGTCCACCGCCGATGATCAACTTCGCTTGCAACCCAGCTTTAGATAAATTAGGCTTTAAGCCAGACCAACGCTTTGCTTATTAA
Protein
MFQTINSKIVLPIIFGVGSVVVLATIIANCLWGKKSKGSSKKLITLVDSNVKYALPLIEREEISHDTRRFRLGLPSPQHVLGLPIGQHIHLSAKINDDLVIRAYTPVSSDEDKGYVDLVIKVYFKNVHPKFPEGGKLSQYLNNMKINDTIDVRGPSGRLQYTGNGTFLIKKLRKDPPTKVVVKKLNLIAGGTGIAPMLQLVRHICTDVNDRTELKLLFANQSEDDILLRDELERYQREHPSQFQVWYTIDRPTDGWKYSSGFINDEMIRDHLFPPSNDVLVLMCGPPPMINFACNPALDKLGFKPDQRFAY
Summary
Catalytic Activity
2 [Fe(III)-cytochrome b5] + NADH = 2 [Fe(II)-cytochrome b5] + H(+) + NAD(+)
Cofactor
FAD
Similarity
Belongs to the flavoprotein pyridine nucleotide cytochrome reductase family.
Uniprot
A0A2A4JIV2
A0A2H1VVY9
S4NZ85
A0A2A4JIP3
A0A3S2L9Y7
A0A2A4JHV5
+ More
U3LUX9 F1DI28 F1CZW3 E2JE74 H9IV72 A0A194R4E9 A0A212F7Z1 A0A194Q8Y9 A0A2P8YM95 A0A0T6AU88 D6WD52 A0A1Y1MT45 J3JV27 A0A1W4WGZ8 A0A1W4WST4 A0A1B0D3G2 A0A1L8DZK0 A0A1L8DZK6 A0A1B0CE42 A0A1A9X321 A0A1L8EGY3 A0A0K8U663 A0A0K8WBM4 A0A1L8ECX1 T1P7R9 A0A1I8MJ03 A0A0B4J2S5 B4QQN4 B4J3C2 B4KYV3 A0A0P8XTI8 A0A1I8P2Z9 A0A0J9UJE3 Q4LDP7 A0A0Q9WS49 B3M4K6 A0A0Q9XDL5 B4N403 A0A232F486 B4HEZ7 Q9I7R1 A0A1B6DUJ7 B4LDM4 D3TNG7 A0A0Q9WKV4 A0A0Q9WJ70 A0A1B6DPJ0 A0A026WBP4 A0A1B0A6S3 A0A336K3J3 A0A0A1WX89 A0A154PE53 B4PFQ1 A0A1A9YKT5 A0A1B0AVZ9 A0A3B0JMM3 A0A1A9URW1 A0A3B0JVI0 A0A3R7M6V6 A0A1B6E148 A0A0R1DX47 A0A0K8TTZ8 A0A0C9S327 A0A1B6EA56 A0A1B6LF45 A0A088AB11 A0A1B6LG70 A0A0R8YFP6 A0A034WPM3 A0A1W4W5E6 A0A034WUU1 A0A1W4VTC1 V9IDL9 E2BQ94 A0A2A3EFN3 V9IBB3 A0A0A9WFH4 A0A1B6CCH9 B4H469 Q29DY6 A0A0M4EK44 A0A0R3P6M7 A0A0A9WQT2 A0A0A9WL13 K1R6L5 W8B1X6 W8BDS6 A0A0K8T4I2 A0A0P4X249 A0A0N7ZDQ1 A0A1B6ENX0 A0A1B6GUP1 A0A0L7QTJ5 U5ET56
U3LUX9 F1DI28 F1CZW3 E2JE74 H9IV72 A0A194R4E9 A0A212F7Z1 A0A194Q8Y9 A0A2P8YM95 A0A0T6AU88 D6WD52 A0A1Y1MT45 J3JV27 A0A1W4WGZ8 A0A1W4WST4 A0A1B0D3G2 A0A1L8DZK0 A0A1L8DZK6 A0A1B0CE42 A0A1A9X321 A0A1L8EGY3 A0A0K8U663 A0A0K8WBM4 A0A1L8ECX1 T1P7R9 A0A1I8MJ03 A0A0B4J2S5 B4QQN4 B4J3C2 B4KYV3 A0A0P8XTI8 A0A1I8P2Z9 A0A0J9UJE3 Q4LDP7 A0A0Q9WS49 B3M4K6 A0A0Q9XDL5 B4N403 A0A232F486 B4HEZ7 Q9I7R1 A0A1B6DUJ7 B4LDM4 D3TNG7 A0A0Q9WKV4 A0A0Q9WJ70 A0A1B6DPJ0 A0A026WBP4 A0A1B0A6S3 A0A336K3J3 A0A0A1WX89 A0A154PE53 B4PFQ1 A0A1A9YKT5 A0A1B0AVZ9 A0A3B0JMM3 A0A1A9URW1 A0A3B0JVI0 A0A3R7M6V6 A0A1B6E148 A0A0R1DX47 A0A0K8TTZ8 A0A0C9S327 A0A1B6EA56 A0A1B6LF45 A0A088AB11 A0A1B6LG70 A0A0R8YFP6 A0A034WPM3 A0A1W4W5E6 A0A034WUU1 A0A1W4VTC1 V9IDL9 E2BQ94 A0A2A3EFN3 V9IBB3 A0A0A9WFH4 A0A1B6CCH9 B4H469 Q29DY6 A0A0M4EK44 A0A0R3P6M7 A0A0A9WQT2 A0A0A9WL13 K1R6L5 W8B1X6 W8BDS6 A0A0K8T4I2 A0A0P4X249 A0A0N7ZDQ1 A0A1B6ENX0 A0A1B6GUP1 A0A0L7QTJ5 U5ET56
EC Number
1.6.2.2
Pubmed
23622113
22986333
19121390
26354079
22118469
29403074
+ More
18362917 19820115 28004739 22516182 25315136 20075255 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20353571 24508170 30249741 25830018 17550304 26369729 25348373 20798317 25401762 26823975 15632085 22992520 24495485
18362917 19820115 28004739 22516182 25315136 20075255 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20353571 24508170 30249741 25830018 17550304 26369729 25348373 20798317 25401762 26823975 15632085 22992520 24495485
EMBL
NWSH01001335
PCG71638.1
ODYU01004755
SOQ44973.1
GAIX01011447
JAA81113.1
+ More
PCG71636.1 RSAL01000074 RVE48925.1 PCG71637.1 JX569756 AGL61416.1 HQ852050 ADX95747.1 HQ638220 ADY89570.1 HQ190046 ADO08221.1 BABH01000621 BABH01000622 BABH01000623 BABH01000624 KQ460761 KPJ12552.1 AGBW02009813 OWR49857.1 KQ459299 KPJ01879.1 PYGN01000495 PSN45371.1 LJIG01022837 KRT78459.1 KQ971312 EEZ98339.1 GEZM01023570 JAV88298.1 BT127093 AEE62055.1 AJVK01023535 AJVK01023536 GFDF01002184 JAV11900.1 GFDF01002183 JAV11901.1 AJWK01008544 GFDG01000885 JAV17914.1 GDHF01033099 GDHF01030268 GDHF01023517 GDHF01017061 JAI19215.1 JAI22046.1 JAI28797.1 JAI35253.1 GDHF01015010 GDHF01008964 GDHF01003716 JAI37304.1 JAI43350.1 JAI48598.1 GFDG01002305 JAV16494.1 KA644666 AFP59295.1 CM000363 CM002912 EDX10136.1 KMY99079.1 CH916366 EDV97221.1 CH933809 EDW18845.1 CH902618 KPU78038.1 KMY99080.1 AE014296 BT082110 AAG22320.1 ACQ45348.1 CH964095 KRF99050.1 EDV39405.1 KRG06389.1 EDW79358.1 NNAY01001033 OXU25412.1 CH480815 EDW41166.1 BT001444 AAF50004.1 AAN71199.1 GEDC01007948 GEDC01002055 JAS29350.1 JAS35243.1 CH940647 EDW69985.1 EZ422969 ADD19245.1 KRF84662.1 KRF84664.1 GEDC01009717 JAS27581.1 KK107292 QOIP01000001 EZA53480.1 RLU27130.1 UFQS01000039 UFQT01000039 SSW98128.1 SSX18514.1 GBXI01010830 GBXI01010398 JAD03462.1 JAD03894.1 KQ434886 KZC10156.1 CM000159 EDW94200.1 JXJN01004436 OUUW01000002 SPP76740.1 SPP76741.1 QCYY01002014 ROT73625.1 GEDC01005642 GEDC01003251 GEDC01000942 GEDC01000865 JAS31656.1 JAS34047.1 JAS36356.1 JAS36433.1 KRK01702.1 GDAI01000168 JAI17435.1 GBYB01015306 JAG85073.1 GEDC01002476 JAS34822.1 GEBQ01017793 JAT22184.1 GEBQ01017281 JAT22696.1 KP083296 ALA55993.1 GAKP01001401 JAC57551.1 GAKP01001407 GAKP01001404 GAKP01001403 GAKP01001402 JAC57548.1 JR038814 AEY58424.1 GL449698 EFN82148.1 KZ288267 PBC30092.1 JR038813 AEY58423.1 GBHO01036407 GDHC01019867 JAG07197.1 JAP98761.1 GEDC01026179 GEDC01020141 GEDC01012611 GEDC01012575 GEDC01006298 JAS11119.1 JAS17157.1 JAS24687.1 JAS24723.1 JAS31000.1 CH479208 EDW31184.1 CH379070 EAL30277.1 CP012525 ALC44476.1 KRT08785.1 GBHO01036405 GBHO01036404 JAG07199.1 JAG07200.1 GBHO01036406 GDHC01021650 GDHC01020694 JAG07198.1 JAP96978.1 JAP97934.1 JH819194 EKC29576.1 GAMC01011415 JAB95140.1 GAMC01011417 GAMC01011414 JAB95138.1 GBRD01005379 JAG60442.1 GDRN01012467 JAI67871.1 GDRN01012466 JAI67872.1 GECZ01030196 JAS39573.1 GECZ01003611 JAS66158.1 KQ414742 KOC61945.1 GANO01002930 JAB56941.1
PCG71636.1 RSAL01000074 RVE48925.1 PCG71637.1 JX569756 AGL61416.1 HQ852050 ADX95747.1 HQ638220 ADY89570.1 HQ190046 ADO08221.1 BABH01000621 BABH01000622 BABH01000623 BABH01000624 KQ460761 KPJ12552.1 AGBW02009813 OWR49857.1 KQ459299 KPJ01879.1 PYGN01000495 PSN45371.1 LJIG01022837 KRT78459.1 KQ971312 EEZ98339.1 GEZM01023570 JAV88298.1 BT127093 AEE62055.1 AJVK01023535 AJVK01023536 GFDF01002184 JAV11900.1 GFDF01002183 JAV11901.1 AJWK01008544 GFDG01000885 JAV17914.1 GDHF01033099 GDHF01030268 GDHF01023517 GDHF01017061 JAI19215.1 JAI22046.1 JAI28797.1 JAI35253.1 GDHF01015010 GDHF01008964 GDHF01003716 JAI37304.1 JAI43350.1 JAI48598.1 GFDG01002305 JAV16494.1 KA644666 AFP59295.1 CM000363 CM002912 EDX10136.1 KMY99079.1 CH916366 EDV97221.1 CH933809 EDW18845.1 CH902618 KPU78038.1 KMY99080.1 AE014296 BT082110 AAG22320.1 ACQ45348.1 CH964095 KRF99050.1 EDV39405.1 KRG06389.1 EDW79358.1 NNAY01001033 OXU25412.1 CH480815 EDW41166.1 BT001444 AAF50004.1 AAN71199.1 GEDC01007948 GEDC01002055 JAS29350.1 JAS35243.1 CH940647 EDW69985.1 EZ422969 ADD19245.1 KRF84662.1 KRF84664.1 GEDC01009717 JAS27581.1 KK107292 QOIP01000001 EZA53480.1 RLU27130.1 UFQS01000039 UFQT01000039 SSW98128.1 SSX18514.1 GBXI01010830 GBXI01010398 JAD03462.1 JAD03894.1 KQ434886 KZC10156.1 CM000159 EDW94200.1 JXJN01004436 OUUW01000002 SPP76740.1 SPP76741.1 QCYY01002014 ROT73625.1 GEDC01005642 GEDC01003251 GEDC01000942 GEDC01000865 JAS31656.1 JAS34047.1 JAS36356.1 JAS36433.1 KRK01702.1 GDAI01000168 JAI17435.1 GBYB01015306 JAG85073.1 GEDC01002476 JAS34822.1 GEBQ01017793 JAT22184.1 GEBQ01017281 JAT22696.1 KP083296 ALA55993.1 GAKP01001401 JAC57551.1 GAKP01001407 GAKP01001404 GAKP01001403 GAKP01001402 JAC57548.1 JR038814 AEY58424.1 GL449698 EFN82148.1 KZ288267 PBC30092.1 JR038813 AEY58423.1 GBHO01036407 GDHC01019867 JAG07197.1 JAP98761.1 GEDC01026179 GEDC01020141 GEDC01012611 GEDC01012575 GEDC01006298 JAS11119.1 JAS17157.1 JAS24687.1 JAS24723.1 JAS31000.1 CH479208 EDW31184.1 CH379070 EAL30277.1 CP012525 ALC44476.1 KRT08785.1 GBHO01036405 GBHO01036404 JAG07199.1 JAG07200.1 GBHO01036406 GDHC01021650 GDHC01020694 JAG07198.1 JAP96978.1 JAP97934.1 JH819194 EKC29576.1 GAMC01011415 JAB95140.1 GAMC01011417 GAMC01011414 JAB95138.1 GBRD01005379 JAG60442.1 GDRN01012467 JAI67871.1 GDRN01012466 JAI67872.1 GECZ01030196 JAS39573.1 GECZ01003611 JAS66158.1 KQ414742 KOC61945.1 GANO01002930 JAB56941.1
Proteomes
UP000218220
UP000283053
UP000005204
UP000053240
UP000007151
UP000053268
+ More
UP000245037 UP000007266 UP000192223 UP000092462 UP000092461 UP000091820 UP000095301 UP000002358 UP000000304 UP000001070 UP000009192 UP000007801 UP000095300 UP000000803 UP000007798 UP000215335 UP000001292 UP000008792 UP000053097 UP000279307 UP000092445 UP000076502 UP000002282 UP000092443 UP000092460 UP000268350 UP000078200 UP000283509 UP000005203 UP000192221 UP000008237 UP000242457 UP000008744 UP000001819 UP000092553 UP000005408 UP000053825
UP000245037 UP000007266 UP000192223 UP000092462 UP000092461 UP000091820 UP000095301 UP000002358 UP000000304 UP000001070 UP000009192 UP000007801 UP000095300 UP000000803 UP000007798 UP000215335 UP000001292 UP000008792 UP000053097 UP000279307 UP000092445 UP000076502 UP000002282 UP000092443 UP000092460 UP000268350 UP000078200 UP000283509 UP000005203 UP000192221 UP000008237 UP000242457 UP000008744 UP000001819 UP000092553 UP000005408 UP000053825
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JIV2
A0A2H1VVY9
S4NZ85
A0A2A4JIP3
A0A3S2L9Y7
A0A2A4JHV5
+ More
U3LUX9 F1DI28 F1CZW3 E2JE74 H9IV72 A0A194R4E9 A0A212F7Z1 A0A194Q8Y9 A0A2P8YM95 A0A0T6AU88 D6WD52 A0A1Y1MT45 J3JV27 A0A1W4WGZ8 A0A1W4WST4 A0A1B0D3G2 A0A1L8DZK0 A0A1L8DZK6 A0A1B0CE42 A0A1A9X321 A0A1L8EGY3 A0A0K8U663 A0A0K8WBM4 A0A1L8ECX1 T1P7R9 A0A1I8MJ03 A0A0B4J2S5 B4QQN4 B4J3C2 B4KYV3 A0A0P8XTI8 A0A1I8P2Z9 A0A0J9UJE3 Q4LDP7 A0A0Q9WS49 B3M4K6 A0A0Q9XDL5 B4N403 A0A232F486 B4HEZ7 Q9I7R1 A0A1B6DUJ7 B4LDM4 D3TNG7 A0A0Q9WKV4 A0A0Q9WJ70 A0A1B6DPJ0 A0A026WBP4 A0A1B0A6S3 A0A336K3J3 A0A0A1WX89 A0A154PE53 B4PFQ1 A0A1A9YKT5 A0A1B0AVZ9 A0A3B0JMM3 A0A1A9URW1 A0A3B0JVI0 A0A3R7M6V6 A0A1B6E148 A0A0R1DX47 A0A0K8TTZ8 A0A0C9S327 A0A1B6EA56 A0A1B6LF45 A0A088AB11 A0A1B6LG70 A0A0R8YFP6 A0A034WPM3 A0A1W4W5E6 A0A034WUU1 A0A1W4VTC1 V9IDL9 E2BQ94 A0A2A3EFN3 V9IBB3 A0A0A9WFH4 A0A1B6CCH9 B4H469 Q29DY6 A0A0M4EK44 A0A0R3P6M7 A0A0A9WQT2 A0A0A9WL13 K1R6L5 W8B1X6 W8BDS6 A0A0K8T4I2 A0A0P4X249 A0A0N7ZDQ1 A0A1B6ENX0 A0A1B6GUP1 A0A0L7QTJ5 U5ET56
U3LUX9 F1DI28 F1CZW3 E2JE74 H9IV72 A0A194R4E9 A0A212F7Z1 A0A194Q8Y9 A0A2P8YM95 A0A0T6AU88 D6WD52 A0A1Y1MT45 J3JV27 A0A1W4WGZ8 A0A1W4WST4 A0A1B0D3G2 A0A1L8DZK0 A0A1L8DZK6 A0A1B0CE42 A0A1A9X321 A0A1L8EGY3 A0A0K8U663 A0A0K8WBM4 A0A1L8ECX1 T1P7R9 A0A1I8MJ03 A0A0B4J2S5 B4QQN4 B4J3C2 B4KYV3 A0A0P8XTI8 A0A1I8P2Z9 A0A0J9UJE3 Q4LDP7 A0A0Q9WS49 B3M4K6 A0A0Q9XDL5 B4N403 A0A232F486 B4HEZ7 Q9I7R1 A0A1B6DUJ7 B4LDM4 D3TNG7 A0A0Q9WKV4 A0A0Q9WJ70 A0A1B6DPJ0 A0A026WBP4 A0A1B0A6S3 A0A336K3J3 A0A0A1WX89 A0A154PE53 B4PFQ1 A0A1A9YKT5 A0A1B0AVZ9 A0A3B0JMM3 A0A1A9URW1 A0A3B0JVI0 A0A3R7M6V6 A0A1B6E148 A0A0R1DX47 A0A0K8TTZ8 A0A0C9S327 A0A1B6EA56 A0A1B6LF45 A0A088AB11 A0A1B6LG70 A0A0R8YFP6 A0A034WPM3 A0A1W4W5E6 A0A034WUU1 A0A1W4VTC1 V9IDL9 E2BQ94 A0A2A3EFN3 V9IBB3 A0A0A9WFH4 A0A1B6CCH9 B4H469 Q29DY6 A0A0M4EK44 A0A0R3P6M7 A0A0A9WQT2 A0A0A9WL13 K1R6L5 W8B1X6 W8BDS6 A0A0K8T4I2 A0A0P4X249 A0A0N7ZDQ1 A0A1B6ENX0 A0A1B6GUP1 A0A0L7QTJ5 U5ET56
PDB
1IB0
E-value=7.21634e-97,
Score=902
Ontologies
KEGG
GO
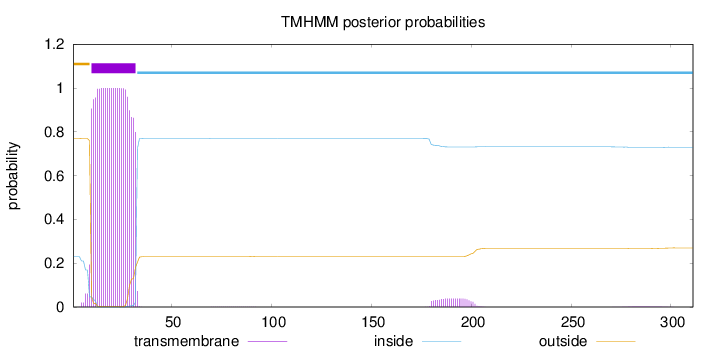
Topology
Length:
311
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.48841
Exp number, first 60 AAs:
22.60846
Total prob of N-in:
0.22930
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 311
Population Genetic Test Statistics
Pi
215.874659
Theta
197.007735
Tajima's D
-1.574005
CLR
16.951684
CSRT
0.0510474476276186
Interpretation
Uncertain
