Gene
KWMTBOMO07965
Pre Gene Modal
BGIBMGA000998
Annotation
PREDICTED:_uncharacterized_protein_LOC106713539_[Papilio_machaon]
Full name
Protein quiver
Alternative Name
Protein sleepless
Location in the cell
Extracellular Reliability : 2.532 PlasmaMembrane Reliability : 1.922
Sequence
CDS
ATGAAGCACTACATTGTATTTTTGTTAATGTTAGGAGCTAATGAATGTTTGGCTACATGGTGCTACCAGTGTACAGCGGCTACTCCTGGTTGTAATGAGCCTTTTAATTGGCGTGGAATGGGATATCTTGGCAATCCATGTCCCGAAAGTGAGGATGTTTGTGTTAAACTTATTGAAAGGAAAGGAGCACAAGAAGTTATTACAAGGGATTGTTTAAGCAATTTTAGAGCTTTCCGAACCGATATTCCTGCTGATACATATGAAGGATGTCGTCCTGCTGCTAAAGATTTAAATCTTGCTCATTATGTCAATAATAGTATAAAAGAGATTGATGTTAAAAGAAATTGGTACGATGAAACCATATGGTGCTTCTGTTTTCTGGACCACCGCTGTAACAATGCATCGAATACATTTGTTTCAATAGCGTTGCTAATGTTATCCGGTGCTACTATATTGTTCAACAAAATATTGTTCTGA
Protein
MKHYIVFLLMLGANECLATWCYQCTAATPGCNEPFNWRGMGYLGNPCPESEDVCVKLIERKGAQEVITRDCLSNFRAFRTDIPADTYEGCRPAAKDLNLAHYVNNSIKEIDVKRNWYDETIWCFCFLDHRCNNASNTFVSIALLMLSGATILFNKILF
Summary
Description
Required for homeostatic regulation of sleep under normal conditions and after sleep deprivation. Important regulator of the Sh K(+) channel, acting as a signaling molecule that connects sleep drive to lowered membrane excitability, possibly by enhancing K(+) channel activity and thus reducing neuronal excitability.
Similarity
Belongs to the quiver family.
Feature
chain Protein quiver
Uniprot
A0A2H1VW45
A0A1E1WTR7
A0A194R415
A0A0L7L088
S4P4R6
A0A194QA60
+ More
A0A212FFF9 A0A2A4JJR0 A0A2M4C1H1 A0A2M4C192 T1E9X4 A0A2M3Z751 A0A2M3ZE51 A0A2M3Z6V8 W5JS25 A0A2M4AY16 A0A2M4AXJ7 A0A2M4AXC9 A0A2M4AWQ3 A0A182Y6X1 A0A182WFP1 A0A0K8VJC5 A0A182WYU6 A0A182FLK1 A0A182P0S7 A0A182QX39 A0A182RLM7 A0A182VG76 A0A182MFG5 A7US48 A0A182HY86 A0A182LHD4 A0A1S4H7A6 A0A1J1I499 A0A182UCL0 A0A034VR30 U5EMH4 A0A3F2YW09 A0A0L0BN68 A0A182GSD6 A0A023EGD3 A0A182GJH4 A0A023EHP3 A0A1S4FYC9 B0W2L7 Q16JT0 A0A1Q3FCC1 A0A182SBF9 A0A182J5S7 B3M4B4 A0A1I8Q615 A0A3B0J4Z9 A0A1A9X0B1 A0A336LKM9 B4N4L5 A0A1B0D3G1 B4HAG8 Q2LZ80 B4KUS8 A0A1I8MCQ4 B4LCR4 T1GHF9 A0A151IUB7 B3NGZ6 A0A1B0CE43 B4PFN2 A0A1L8D9K6 A0A151IN56 A0A1B0AEH0 A0A1B0BY91 B4IZB7 A0A1W4X4S8 A0A0M4F1Z0 A0A195F7I1 A0A1B0GBB9 A0A158NYC9 A0A1A9VP42 A0A3L8DXU7 A0A0L7RHA1 A0A2P8YMD6 B4QQA2 R4G374 Q9VTM6 A0A151X886 B4HEX7 A0A232EYU4 A0A1B6CQR7 A0A1W4VVF9 E2A4B4 A0A154PFI8 A0A026X3L7 A0A067RGR4 E2BDF0 K7IZ12 A0A1B6IIT2 A0A1B6MTQ1 A0A1B6GJT0 D6WAX2 A0A310S7H4
A0A212FFF9 A0A2A4JJR0 A0A2M4C1H1 A0A2M4C192 T1E9X4 A0A2M3Z751 A0A2M3ZE51 A0A2M3Z6V8 W5JS25 A0A2M4AY16 A0A2M4AXJ7 A0A2M4AXC9 A0A2M4AWQ3 A0A182Y6X1 A0A182WFP1 A0A0K8VJC5 A0A182WYU6 A0A182FLK1 A0A182P0S7 A0A182QX39 A0A182RLM7 A0A182VG76 A0A182MFG5 A7US48 A0A182HY86 A0A182LHD4 A0A1S4H7A6 A0A1J1I499 A0A182UCL0 A0A034VR30 U5EMH4 A0A3F2YW09 A0A0L0BN68 A0A182GSD6 A0A023EGD3 A0A182GJH4 A0A023EHP3 A0A1S4FYC9 B0W2L7 Q16JT0 A0A1Q3FCC1 A0A182SBF9 A0A182J5S7 B3M4B4 A0A1I8Q615 A0A3B0J4Z9 A0A1A9X0B1 A0A336LKM9 B4N4L5 A0A1B0D3G1 B4HAG8 Q2LZ80 B4KUS8 A0A1I8MCQ4 B4LCR4 T1GHF9 A0A151IUB7 B3NGZ6 A0A1B0CE43 B4PFN2 A0A1L8D9K6 A0A151IN56 A0A1B0AEH0 A0A1B0BY91 B4IZB7 A0A1W4X4S8 A0A0M4F1Z0 A0A195F7I1 A0A1B0GBB9 A0A158NYC9 A0A1A9VP42 A0A3L8DXU7 A0A0L7RHA1 A0A2P8YMD6 B4QQA2 R4G374 Q9VTM6 A0A151X886 B4HEX7 A0A232EYU4 A0A1B6CQR7 A0A1W4VVF9 E2A4B4 A0A154PFI8 A0A026X3L7 A0A067RGR4 E2BDF0 K7IZ12 A0A1B6IIT2 A0A1B6MTQ1 A0A1B6GJT0 D6WAX2 A0A310S7H4
Pubmed
26354079
26227816
23622113
22118469
20920257
23761445
+ More
25244985 12364791 20966253 25348373 26108605 26483478 24945155 17510324 17994087 15632085 25315136 17550304 21347285 30249741 29403074 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20798317 24508170 24845553 20075255 18362917 19820115
25244985 12364791 20966253 25348373 26108605 26483478 24945155 17510324 17994087 15632085 25315136 17550304 21347285 30249741 29403074 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20798317 24508170 24845553 20075255 18362917 19820115
EMBL
ODYU01004755
SOQ44976.1
GDQN01000853
JAT90201.1
KQ460761
KPJ12553.1
+ More
JTDY01004036 KOB68684.1 GAIX01011085 JAA81475.1 KQ459299 KPJ01880.1 AGBW02008828 OWR52471.1 NWSH01001335 PCG71640.1 GGFJ01010008 MBW59149.1 GGFJ01009955 MBW59096.1 GAMD01000922 JAB00669.1 GGFM01003596 MBW24347.1 GGFM01006041 MBW26792.1 GGFM01003490 MBW24241.1 ADMH02000630 ETN65549.1 GGFK01012281 MBW45602.1 GGFK01012111 MBW45432.1 GGFK01012138 MBW45459.1 GGFK01011893 MBW45214.1 GDHF01013318 JAI38996.1 AXCN02001110 AXCM01003816 AAAB01008816 EDO64459.1 APCN01004507 APCN01004508 CVRI01000037 CRK93694.1 GAKP01014018 JAC44934.1 GANO01004477 JAB55394.1 JRES01001603 KNC21555.1 JXUM01084469 KQ563439 KXJ73818.1 GAPW01005192 GAPW01005191 JAC08407.1 JXUM01068210 KQ562488 KXJ75799.1 GAPW01005193 JAC08405.1 DS231827 EDS29325.1 CH477993 EAT34553.1 GFDL01009838 JAV25207.1 CH902618 EDV39384.1 OUUW01000002 SPP76767.1 UFQT01000039 SSX18516.1 CH964095 EDW79089.1 AJVK01023535 CH479240 EDW37566.1 CH379069 EAL29628.2 CH933809 EDW19334.1 CH940647 EDW70956.1 CAQQ02197562 CAQQ02197563 KQ980963 KYN11013.1 CH954178 EDV51453.1 AJWK01008544 CM000159 EDW94181.1 GFDF01011047 JAV03037.1 KQ976963 KYN06779.1 JXJN01022542 CH916366 EDV96672.1 CP012525 ALC45057.1 KQ981768 KYN36024.1 CCAG010022534 ADTU01003755 QOIP01000003 RLU25063.1 KQ414591 KOC70250.1 PYGN01000495 PSN45393.1 CM000363 CM002912 EDX10120.1 KMY99056.1 GAHY01001936 JAA75574.1 AE014296 AY060768 KX531850 AAF50021.1 AAL28316.1 ANY27660.1 KQ982426 KYQ56582.1 CH480815 EDW41146.1 NNAY01001559 OXU23602.1 GEDC01021516 JAS15782.1 GL436621 EFN71724.1 KQ434879 KZC10000.1 KK107013 EZA62867.1 KK852480 KDR22967.1 GL447620 EFN86262.1 GECU01020872 JAS86834.1 GEBQ01000718 JAT39259.1 GECZ01029976 GECZ01007216 GECZ01001445 JAS39793.1 JAS62553.1 JAS68324.1 KQ971312 EEZ97935.1 KQ765809 OAD53797.1
JTDY01004036 KOB68684.1 GAIX01011085 JAA81475.1 KQ459299 KPJ01880.1 AGBW02008828 OWR52471.1 NWSH01001335 PCG71640.1 GGFJ01010008 MBW59149.1 GGFJ01009955 MBW59096.1 GAMD01000922 JAB00669.1 GGFM01003596 MBW24347.1 GGFM01006041 MBW26792.1 GGFM01003490 MBW24241.1 ADMH02000630 ETN65549.1 GGFK01012281 MBW45602.1 GGFK01012111 MBW45432.1 GGFK01012138 MBW45459.1 GGFK01011893 MBW45214.1 GDHF01013318 JAI38996.1 AXCN02001110 AXCM01003816 AAAB01008816 EDO64459.1 APCN01004507 APCN01004508 CVRI01000037 CRK93694.1 GAKP01014018 JAC44934.1 GANO01004477 JAB55394.1 JRES01001603 KNC21555.1 JXUM01084469 KQ563439 KXJ73818.1 GAPW01005192 GAPW01005191 JAC08407.1 JXUM01068210 KQ562488 KXJ75799.1 GAPW01005193 JAC08405.1 DS231827 EDS29325.1 CH477993 EAT34553.1 GFDL01009838 JAV25207.1 CH902618 EDV39384.1 OUUW01000002 SPP76767.1 UFQT01000039 SSX18516.1 CH964095 EDW79089.1 AJVK01023535 CH479240 EDW37566.1 CH379069 EAL29628.2 CH933809 EDW19334.1 CH940647 EDW70956.1 CAQQ02197562 CAQQ02197563 KQ980963 KYN11013.1 CH954178 EDV51453.1 AJWK01008544 CM000159 EDW94181.1 GFDF01011047 JAV03037.1 KQ976963 KYN06779.1 JXJN01022542 CH916366 EDV96672.1 CP012525 ALC45057.1 KQ981768 KYN36024.1 CCAG010022534 ADTU01003755 QOIP01000003 RLU25063.1 KQ414591 KOC70250.1 PYGN01000495 PSN45393.1 CM000363 CM002912 EDX10120.1 KMY99056.1 GAHY01001936 JAA75574.1 AE014296 AY060768 KX531850 AAF50021.1 AAL28316.1 ANY27660.1 KQ982426 KYQ56582.1 CH480815 EDW41146.1 NNAY01001559 OXU23602.1 GEDC01021516 JAS15782.1 GL436621 EFN71724.1 KQ434879 KZC10000.1 KK107013 EZA62867.1 KK852480 KDR22967.1 GL447620 EFN86262.1 GECU01020872 JAS86834.1 GEBQ01000718 JAT39259.1 GECZ01029976 GECZ01007216 GECZ01001445 JAS39793.1 JAS62553.1 JAS68324.1 KQ971312 EEZ97935.1 KQ765809 OAD53797.1
Proteomes
UP000053240
UP000037510
UP000053268
UP000007151
UP000218220
UP000000673
+ More
UP000076408 UP000075920 UP000076407 UP000069272 UP000075885 UP000075886 UP000075900 UP000075903 UP000075883 UP000007062 UP000075840 UP000075882 UP000183832 UP000075902 UP000075884 UP000037069 UP000069940 UP000249989 UP000002320 UP000008820 UP000075901 UP000075880 UP000007801 UP000095300 UP000268350 UP000091820 UP000007798 UP000092462 UP000008744 UP000001819 UP000009192 UP000095301 UP000008792 UP000015102 UP000078492 UP000008711 UP000092461 UP000002282 UP000078542 UP000092445 UP000092460 UP000001070 UP000192223 UP000092553 UP000078541 UP000092444 UP000005205 UP000078200 UP000279307 UP000053825 UP000245037 UP000000304 UP000000803 UP000075809 UP000001292 UP000215335 UP000192221 UP000000311 UP000076502 UP000053097 UP000027135 UP000008237 UP000002358 UP000007266
UP000076408 UP000075920 UP000076407 UP000069272 UP000075885 UP000075886 UP000075900 UP000075903 UP000075883 UP000007062 UP000075840 UP000075882 UP000183832 UP000075902 UP000075884 UP000037069 UP000069940 UP000249989 UP000002320 UP000008820 UP000075901 UP000075880 UP000007801 UP000095300 UP000268350 UP000091820 UP000007798 UP000092462 UP000008744 UP000001819 UP000009192 UP000095301 UP000008792 UP000015102 UP000078492 UP000008711 UP000092461 UP000002282 UP000078542 UP000092445 UP000092460 UP000001070 UP000192223 UP000092553 UP000078541 UP000092444 UP000005205 UP000078200 UP000279307 UP000053825 UP000245037 UP000000304 UP000000803 UP000075809 UP000001292 UP000215335 UP000192221 UP000000311 UP000076502 UP000053097 UP000027135 UP000008237 UP000002358 UP000007266
PRIDE
ProteinModelPortal
A0A2H1VW45
A0A1E1WTR7
A0A194R415
A0A0L7L088
S4P4R6
A0A194QA60
+ More
A0A212FFF9 A0A2A4JJR0 A0A2M4C1H1 A0A2M4C192 T1E9X4 A0A2M3Z751 A0A2M3ZE51 A0A2M3Z6V8 W5JS25 A0A2M4AY16 A0A2M4AXJ7 A0A2M4AXC9 A0A2M4AWQ3 A0A182Y6X1 A0A182WFP1 A0A0K8VJC5 A0A182WYU6 A0A182FLK1 A0A182P0S7 A0A182QX39 A0A182RLM7 A0A182VG76 A0A182MFG5 A7US48 A0A182HY86 A0A182LHD4 A0A1S4H7A6 A0A1J1I499 A0A182UCL0 A0A034VR30 U5EMH4 A0A3F2YW09 A0A0L0BN68 A0A182GSD6 A0A023EGD3 A0A182GJH4 A0A023EHP3 A0A1S4FYC9 B0W2L7 Q16JT0 A0A1Q3FCC1 A0A182SBF9 A0A182J5S7 B3M4B4 A0A1I8Q615 A0A3B0J4Z9 A0A1A9X0B1 A0A336LKM9 B4N4L5 A0A1B0D3G1 B4HAG8 Q2LZ80 B4KUS8 A0A1I8MCQ4 B4LCR4 T1GHF9 A0A151IUB7 B3NGZ6 A0A1B0CE43 B4PFN2 A0A1L8D9K6 A0A151IN56 A0A1B0AEH0 A0A1B0BY91 B4IZB7 A0A1W4X4S8 A0A0M4F1Z0 A0A195F7I1 A0A1B0GBB9 A0A158NYC9 A0A1A9VP42 A0A3L8DXU7 A0A0L7RHA1 A0A2P8YMD6 B4QQA2 R4G374 Q9VTM6 A0A151X886 B4HEX7 A0A232EYU4 A0A1B6CQR7 A0A1W4VVF9 E2A4B4 A0A154PFI8 A0A026X3L7 A0A067RGR4 E2BDF0 K7IZ12 A0A1B6IIT2 A0A1B6MTQ1 A0A1B6GJT0 D6WAX2 A0A310S7H4
A0A212FFF9 A0A2A4JJR0 A0A2M4C1H1 A0A2M4C192 T1E9X4 A0A2M3Z751 A0A2M3ZE51 A0A2M3Z6V8 W5JS25 A0A2M4AY16 A0A2M4AXJ7 A0A2M4AXC9 A0A2M4AWQ3 A0A182Y6X1 A0A182WFP1 A0A0K8VJC5 A0A182WYU6 A0A182FLK1 A0A182P0S7 A0A182QX39 A0A182RLM7 A0A182VG76 A0A182MFG5 A7US48 A0A182HY86 A0A182LHD4 A0A1S4H7A6 A0A1J1I499 A0A182UCL0 A0A034VR30 U5EMH4 A0A3F2YW09 A0A0L0BN68 A0A182GSD6 A0A023EGD3 A0A182GJH4 A0A023EHP3 A0A1S4FYC9 B0W2L7 Q16JT0 A0A1Q3FCC1 A0A182SBF9 A0A182J5S7 B3M4B4 A0A1I8Q615 A0A3B0J4Z9 A0A1A9X0B1 A0A336LKM9 B4N4L5 A0A1B0D3G1 B4HAG8 Q2LZ80 B4KUS8 A0A1I8MCQ4 B4LCR4 T1GHF9 A0A151IUB7 B3NGZ6 A0A1B0CE43 B4PFN2 A0A1L8D9K6 A0A151IN56 A0A1B0AEH0 A0A1B0BY91 B4IZB7 A0A1W4X4S8 A0A0M4F1Z0 A0A195F7I1 A0A1B0GBB9 A0A158NYC9 A0A1A9VP42 A0A3L8DXU7 A0A0L7RHA1 A0A2P8YMD6 B4QQA2 R4G374 Q9VTM6 A0A151X886 B4HEX7 A0A232EYU4 A0A1B6CQR7 A0A1W4VVF9 E2A4B4 A0A154PFI8 A0A026X3L7 A0A067RGR4 E2BDF0 K7IZ12 A0A1B6IIT2 A0A1B6MTQ1 A0A1B6GJT0 D6WAX2 A0A310S7H4
Ontologies
GO
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 18,
Likelihood: 0.999429
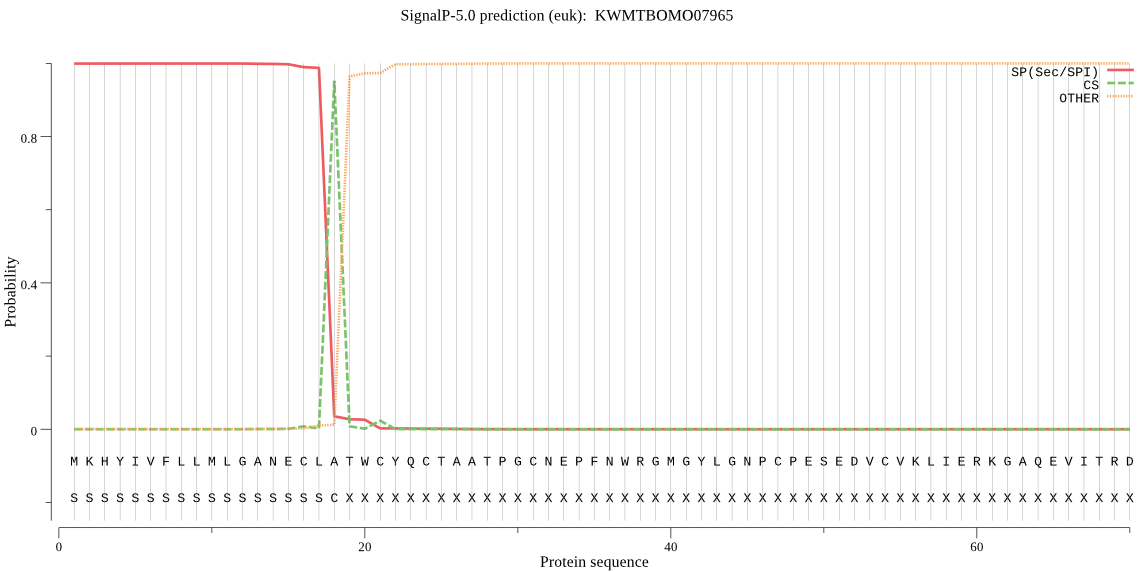
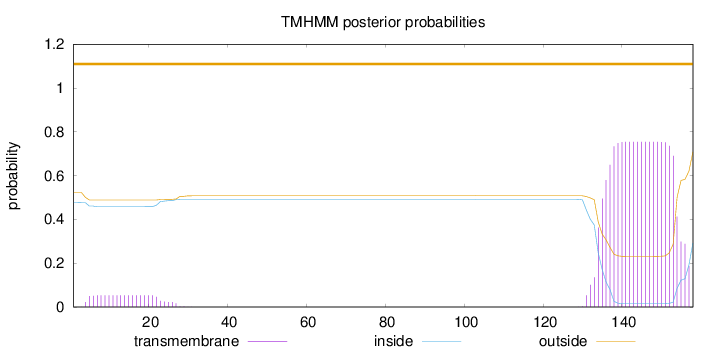
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
16.61393
Exp number, first 60 AAs:
1.09461
Total prob of N-in:
0.47696
outside
1 - 158
Population Genetic Test Statistics
Pi
128.490717
Theta
136.659105
Tajima's D
-0.37175
CLR
0.307945
CSRT
0.268986550672466
Interpretation
Uncertain
