Gene
KWMTBOMO07962
Pre Gene Modal
BGIBMGA000996
Annotation
PREDICTED:_uncharacterized_protein_LOC101746483_isoform_X2_[Bombyx_mori]
Full name
Protein quiver
+ More
Multifunctional fusion protein
Multifunctional fusion protein
Alternative Name
Protein sleepless
Location in the cell
Extracellular Reliability : 2.458 PlasmaMembrane Reliability : 1.971
Sequence
CDS
ATGAAACTCGAAGTATTGTTGACAACTGTGCTAGCGATAAGTTACCTTAAAGTTGAAATCAGCTGTTTAAGTAGGAGATGTATACAGTGTAGATCTCGAGGAGAGCTTGGGTCATGTGGAGACCCTCTACCCTTTAATATAAGTGACCCTGAAGCAGAACATGGTGTGCATATTACTGCTTGTCCATCTGGATGGTGTGCCAAGAGAATTCAGGGAACCACTGGCACATTTAGAACTGATGATTATGGTGCGGTAACAGAAAGGTCCTGTCTTCAACAGCCTCCTAGTGATTATGAAGAAAGATGTGCGTATACCATGTGGAAATATAAAAGGGTATATGTATGTTTCTGTAATGGTGATTTGTGCAATTCAGCTCCAACAATGAAGATGTTTTCTCCTGTATTATTGCTCAGTATTTTTATCTTCATAAAGAATTATATTTATTAA
Protein
MKLEVLLTTVLAISYLKVEISCLSRRCIQCRSRGELGSCGDPLPFNISDPEAEHGVHITACPSGWCAKRIQGTTGTFRTDDYGAVTERSCLQQPPSDYEERCAYTMWKYKRVYVCFCNGDLCNSAPTMKMFSPVLLLSIFIFIKNYIY
Summary
Description
Required for homeostatic regulation of sleep under normal conditions and after sleep deprivation. Important regulator of the Sh K(+) channel, acting as a signaling molecule that connects sleep drive to lowered membrane excitability, possibly by enhancing K(+) channel activity and thus reducing neuronal excitability.
Stores iron in a soluble, non-toxic, readily available form. Important for iron homeostasis. Iron is taken up in the ferrous form and deposited as ferric hydroxides after oxidation.
Stores iron in a soluble, non-toxic, readily available form. Important for iron homeostasis. Iron is taken up in the ferrous form and deposited as ferric hydroxides after oxidation.
Catalytic Activity
4 Fe(2+) + 4 H(+) + O2 = 4 Fe(3+) + 2 H2O
Similarity
Belongs to the quiver family.
Belongs to the ferritin family.
Belongs to the ferritin family.
Feature
chain Protein quiver
Uniprot
H9IUR6
A0A194R9M4
S4P4T8
A0A3S2M1C8
A0A1E1WN51
I4DKP0
+ More
I4DKN9 A0A2A4JXT4 A0A2H1WLM9 A0A182H2X5 A0A023EDS6 A0A2M4AWN9 A0A2M4CJZ8 A0A2M3Z3A8 A0A2M4AWT0 A0A1S4F1U2 Q17I69 A0A2M4C2T7 B0X179 A0A1I8NDP3 A0A182WR58 A0A182LPP4 Q7Q2Z7 A0A182HNQ1 A0A0M5JDS4 A0A182VJC0 A0A182FJA9 A0A2J7PYC1 A0A067QST0 A0A1L8DP67 A0A2M4C335 A0A182MZT5 B4F5Y8 A0A182G608 B4F5Z0 Q9VZ21 B4IDZ0 B4NUA5 A0A0L0C0B6 A0A2M4AXJ8 A0A182RVP6 B4F5Z6 A0A182YH23 A0A182W5Y2 B4F5Z5 A0A182U521 B4PYE6 B4F5Z3 B3MVY1 A0A1J1ISB0 A0A0C9QQ49 A0A1W4V1L9 A0A182MST6 B4NCI9 A0A1I8PPE1 A0A2R7VU52 A0A224XPJ4 A0A161MT98 A0A182QS10 A0A084VDE1 A0A182K5P4 A0A3B0JWZ6 A0A088AE79 A0A1Y1NPU0 A0A1A9X3A5 T1ICK5 T1GCM6 A0A1W4XEN6 B4K2D2 B4M2N1 B4JJL3 A0A1A9YDD2 A0A1B0B035 Q29HJ3 A0A195FEF0 B4L4H7 A0A1B6INM5 J9ZZN6 A0A1B0G421 A0A1B0A3H8 A0A195BG51 A0A1A9VC03 A0A195CBF6 A0A1B6DF33 A0A336LRB1 A0A1B6FII3 B4GXK7 A0A2P8XRD8 A0A1S3CXZ1 B3NV97 A0A151J979 A0A0A9XAT5 C4WX31 E0VCD4 A0A2A3ERS0 E2BID2 A0A226CXF1 A0A226E6J0 A0A0T6B6S7 A0A0N0BFP8
I4DKN9 A0A2A4JXT4 A0A2H1WLM9 A0A182H2X5 A0A023EDS6 A0A2M4AWN9 A0A2M4CJZ8 A0A2M3Z3A8 A0A2M4AWT0 A0A1S4F1U2 Q17I69 A0A2M4C2T7 B0X179 A0A1I8NDP3 A0A182WR58 A0A182LPP4 Q7Q2Z7 A0A182HNQ1 A0A0M5JDS4 A0A182VJC0 A0A182FJA9 A0A2J7PYC1 A0A067QST0 A0A1L8DP67 A0A2M4C335 A0A182MZT5 B4F5Y8 A0A182G608 B4F5Z0 Q9VZ21 B4IDZ0 B4NUA5 A0A0L0C0B6 A0A2M4AXJ8 A0A182RVP6 B4F5Z6 A0A182YH23 A0A182W5Y2 B4F5Z5 A0A182U521 B4PYE6 B4F5Z3 B3MVY1 A0A1J1ISB0 A0A0C9QQ49 A0A1W4V1L9 A0A182MST6 B4NCI9 A0A1I8PPE1 A0A2R7VU52 A0A224XPJ4 A0A161MT98 A0A182QS10 A0A084VDE1 A0A182K5P4 A0A3B0JWZ6 A0A088AE79 A0A1Y1NPU0 A0A1A9X3A5 T1ICK5 T1GCM6 A0A1W4XEN6 B4K2D2 B4M2N1 B4JJL3 A0A1A9YDD2 A0A1B0B035 Q29HJ3 A0A195FEF0 B4L4H7 A0A1B6INM5 J9ZZN6 A0A1B0G421 A0A1B0A3H8 A0A195BG51 A0A1A9VC03 A0A195CBF6 A0A1B6DF33 A0A336LRB1 A0A1B6FII3 B4GXK7 A0A2P8XRD8 A0A1S3CXZ1 B3NV97 A0A151J979 A0A0A9XAT5 C4WX31 E0VCD4 A0A2A3ERS0 E2BID2 A0A226CXF1 A0A226E6J0 A0A0T6B6S7 A0A0N0BFP8
Pubmed
19121390
26354079
23622113
22651552
26483478
24945155
+ More
17510324 25315136 20966253 12364791 14747013 17210077 24845553 18477586 19126864 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 26108605 25244985 17550304 24438588 28004739 15632085 18362917 19820115 29403074 25401762 20566863 20798317
17510324 25315136 20966253 12364791 14747013 17210077 24845553 18477586 19126864 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 26108605 25244985 17550304 24438588 28004739 15632085 18362917 19820115 29403074 25401762 20566863 20798317
EMBL
BABH01000633
KQ460761
KPJ12556.1
GAIX01007516
JAA85044.1
RSAL01000074
+ More
RVE48921.1 GDQN01002595 JAT88459.1 AK401858 KQ459299 BAM18480.1 KPJ01883.1 AK401857 BAM18479.1 NWSH01000379 PCG76831.1 ODYU01009497 SOQ53981.1 JXUM01106183 JXUM01106184 JXUM01106185 KQ565031 KXJ71423.1 GAPW01005860 JAC07738.1 GGFK01011896 MBW45217.1 GGFL01001387 MBW65565.1 GGFM01002256 MBW23007.1 GGFK01011894 MBW45215.1 CH477242 EAT46360.1 GGFJ01010177 MBW59318.1 DS232255 EDS38528.1 AAAB01008966 EAA13014.4 APCN01001962 CP012528 ALC49306.1 NEVH01020850 PNF21312.1 KK853324 KDR08506.1 GFDF01005939 JAV08145.1 GGFJ01010544 MBW59685.1 AM999224 AM999225 AM999227 AM999228 AM999230 FM246342 FM246346 FM246347 FM246348 FM246349 CAQ53586.1 CAQ53587.1 CAQ53589.1 CAQ53590.1 CAQ53592.1 CAR94268.1 CAR94272.1 CAR94273.1 CAR94274.1 CAR94275.1 JXUM01007400 JXUM01007401 KQ560239 KXJ83657.1 AM999226 CAQ53588.1 AE014298 AY061126 KX531353 AM999233 AM999234 AM999235 FM246338 FM246339 FM246340 FM246341 FM246343 FM246344 FM246345 AAF48010.1 AAL28674.1 ANY27163.1 CAQ53595.1 CAQ53596.1 CAQ53597.1 CAR94264.1 CAR94265.1 CAR94266.1 CAR94267.1 CAR94269.1 CAR94270.1 CAR94271.1 CH480830 EDW45798.1 CH983813 EDX16552.1 JRES01001072 KNC25733.1 GGFK01012205 MBW45526.1 AM999232 CAQ53594.1 AM999231 CAQ53593.1 CM000162 EDX02008.1 AM999229 CAQ53591.1 CH902625 EDV35126.1 CVRI01000057 CRL01998.1 GBYB01005799 JAG75566.1 AXCM01011918 CH964239 EDW82548.2 KK854082 PTY10889.1 GFTR01002020 JAW14406.1 GEMB01000256 JAS02859.1 AXCN02001405 ATLV01011272 KE524660 KFB35985.1 OUUW01000011 SPP86574.1 GEZM01000724 JAV98297.1 ACPB03022220 CAQQ02082737 CH918959 EDW04389.1 CH940651 EDW65935.1 CH916370 EDV99765.1 JXJN01006551 CH379064 EAL31765.3 KQ981673 KYN38399.1 CH933810 EDW07455.1 GECU01019178 JAS88528.1 JX470185 KQ971338 AFS64713.1 KYB27965.1 CCAG010020282 KQ976500 KYM83157.1 KQ978068 KYM97536.1 GEDC01012990 JAS24308.1 UFQS01000116 UFQT01000116 SSW99945.1 SSX20325.1 GECZ01019757 JAS50012.1 CH479196 EDW27484.1 PYGN01001493 PSN34474.1 CH954180 EDV46085.1 KQ979433 KYN21571.1 GBHO01027676 GBHO01027675 JAG15928.1 JAG15929.1 ABLF02029452 AK342418 BAH72451.1 DS235053 EEB11040.1 KZ288192 PBC34390.1 GL448504 EFN84520.1 LNIX01000077 OXA36716.1 LNIX01000006 OXA53232.1 LJIG01009450 KRT83055.1 KQ435794 KOX73661.1
RVE48921.1 GDQN01002595 JAT88459.1 AK401858 KQ459299 BAM18480.1 KPJ01883.1 AK401857 BAM18479.1 NWSH01000379 PCG76831.1 ODYU01009497 SOQ53981.1 JXUM01106183 JXUM01106184 JXUM01106185 KQ565031 KXJ71423.1 GAPW01005860 JAC07738.1 GGFK01011896 MBW45217.1 GGFL01001387 MBW65565.1 GGFM01002256 MBW23007.1 GGFK01011894 MBW45215.1 CH477242 EAT46360.1 GGFJ01010177 MBW59318.1 DS232255 EDS38528.1 AAAB01008966 EAA13014.4 APCN01001962 CP012528 ALC49306.1 NEVH01020850 PNF21312.1 KK853324 KDR08506.1 GFDF01005939 JAV08145.1 GGFJ01010544 MBW59685.1 AM999224 AM999225 AM999227 AM999228 AM999230 FM246342 FM246346 FM246347 FM246348 FM246349 CAQ53586.1 CAQ53587.1 CAQ53589.1 CAQ53590.1 CAQ53592.1 CAR94268.1 CAR94272.1 CAR94273.1 CAR94274.1 CAR94275.1 JXUM01007400 JXUM01007401 KQ560239 KXJ83657.1 AM999226 CAQ53588.1 AE014298 AY061126 KX531353 AM999233 AM999234 AM999235 FM246338 FM246339 FM246340 FM246341 FM246343 FM246344 FM246345 AAF48010.1 AAL28674.1 ANY27163.1 CAQ53595.1 CAQ53596.1 CAQ53597.1 CAR94264.1 CAR94265.1 CAR94266.1 CAR94267.1 CAR94269.1 CAR94270.1 CAR94271.1 CH480830 EDW45798.1 CH983813 EDX16552.1 JRES01001072 KNC25733.1 GGFK01012205 MBW45526.1 AM999232 CAQ53594.1 AM999231 CAQ53593.1 CM000162 EDX02008.1 AM999229 CAQ53591.1 CH902625 EDV35126.1 CVRI01000057 CRL01998.1 GBYB01005799 JAG75566.1 AXCM01011918 CH964239 EDW82548.2 KK854082 PTY10889.1 GFTR01002020 JAW14406.1 GEMB01000256 JAS02859.1 AXCN02001405 ATLV01011272 KE524660 KFB35985.1 OUUW01000011 SPP86574.1 GEZM01000724 JAV98297.1 ACPB03022220 CAQQ02082737 CH918959 EDW04389.1 CH940651 EDW65935.1 CH916370 EDV99765.1 JXJN01006551 CH379064 EAL31765.3 KQ981673 KYN38399.1 CH933810 EDW07455.1 GECU01019178 JAS88528.1 JX470185 KQ971338 AFS64713.1 KYB27965.1 CCAG010020282 KQ976500 KYM83157.1 KQ978068 KYM97536.1 GEDC01012990 JAS24308.1 UFQS01000116 UFQT01000116 SSW99945.1 SSX20325.1 GECZ01019757 JAS50012.1 CH479196 EDW27484.1 PYGN01001493 PSN34474.1 CH954180 EDV46085.1 KQ979433 KYN21571.1 GBHO01027676 GBHO01027675 JAG15928.1 JAG15929.1 ABLF02029452 AK342418 BAH72451.1 DS235053 EEB11040.1 KZ288192 PBC34390.1 GL448504 EFN84520.1 LNIX01000077 OXA36716.1 LNIX01000006 OXA53232.1 LJIG01009450 KRT83055.1 KQ435794 KOX73661.1
Proteomes
UP000005204
UP000053240
UP000283053
UP000053268
UP000218220
UP000069940
+ More
UP000249989 UP000008820 UP000002320 UP000095301 UP000076407 UP000075882 UP000007062 UP000075840 UP000092553 UP000075903 UP000069272 UP000235965 UP000027135 UP000075884 UP000000803 UP000001292 UP000000304 UP000037069 UP000075900 UP000076408 UP000075920 UP000075902 UP000002282 UP000007801 UP000183832 UP000192221 UP000075883 UP000007798 UP000095300 UP000075886 UP000030765 UP000075881 UP000268350 UP000005203 UP000091820 UP000015103 UP000015102 UP000192223 UP000001070 UP000008792 UP000092443 UP000092460 UP000001819 UP000078541 UP000009192 UP000007266 UP000092444 UP000092445 UP000078540 UP000078200 UP000078542 UP000008744 UP000245037 UP000079169 UP000008711 UP000078492 UP000007819 UP000009046 UP000242457 UP000008237 UP000198287 UP000053105
UP000249989 UP000008820 UP000002320 UP000095301 UP000076407 UP000075882 UP000007062 UP000075840 UP000092553 UP000075903 UP000069272 UP000235965 UP000027135 UP000075884 UP000000803 UP000001292 UP000000304 UP000037069 UP000075900 UP000076408 UP000075920 UP000075902 UP000002282 UP000007801 UP000183832 UP000192221 UP000075883 UP000007798 UP000095300 UP000075886 UP000030765 UP000075881 UP000268350 UP000005203 UP000091820 UP000015103 UP000015102 UP000192223 UP000001070 UP000008792 UP000092443 UP000092460 UP000001819 UP000078541 UP000009192 UP000007266 UP000092444 UP000092445 UP000078540 UP000078200 UP000078542 UP000008744 UP000245037 UP000079169 UP000008711 UP000078492 UP000007819 UP000009046 UP000242457 UP000008237 UP000198287 UP000053105
Interpro
IPR031424
QVR
+ More
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR014034 Ferritin_CS
IPR009040 Ferritin-like_diiron
IPR008331 Ferritin_DPS_dom
IPR009078 Ferritin-like_SF
IPR012347 Ferritin-like
IPR001519 Ferritin
IPR038905 ARMC2
IPR036179 Ig-like_dom_sf
IPR013151 Immunoglobulin
IPR013783 Ig-like_fold
IPR039311 FAM187A/B
IPR007110 Ig-like_dom
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR014034 Ferritin_CS
IPR009040 Ferritin-like_diiron
IPR008331 Ferritin_DPS_dom
IPR009078 Ferritin-like_SF
IPR012347 Ferritin-like
IPR001519 Ferritin
IPR038905 ARMC2
IPR036179 Ig-like_dom_sf
IPR013151 Immunoglobulin
IPR013783 Ig-like_fold
IPR039311 FAM187A/B
IPR007110 Ig-like_dom
Gene 3D
ProteinModelPortal
H9IUR6
A0A194R9M4
S4P4T8
A0A3S2M1C8
A0A1E1WN51
I4DKP0
+ More
I4DKN9 A0A2A4JXT4 A0A2H1WLM9 A0A182H2X5 A0A023EDS6 A0A2M4AWN9 A0A2M4CJZ8 A0A2M3Z3A8 A0A2M4AWT0 A0A1S4F1U2 Q17I69 A0A2M4C2T7 B0X179 A0A1I8NDP3 A0A182WR58 A0A182LPP4 Q7Q2Z7 A0A182HNQ1 A0A0M5JDS4 A0A182VJC0 A0A182FJA9 A0A2J7PYC1 A0A067QST0 A0A1L8DP67 A0A2M4C335 A0A182MZT5 B4F5Y8 A0A182G608 B4F5Z0 Q9VZ21 B4IDZ0 B4NUA5 A0A0L0C0B6 A0A2M4AXJ8 A0A182RVP6 B4F5Z6 A0A182YH23 A0A182W5Y2 B4F5Z5 A0A182U521 B4PYE6 B4F5Z3 B3MVY1 A0A1J1ISB0 A0A0C9QQ49 A0A1W4V1L9 A0A182MST6 B4NCI9 A0A1I8PPE1 A0A2R7VU52 A0A224XPJ4 A0A161MT98 A0A182QS10 A0A084VDE1 A0A182K5P4 A0A3B0JWZ6 A0A088AE79 A0A1Y1NPU0 A0A1A9X3A5 T1ICK5 T1GCM6 A0A1W4XEN6 B4K2D2 B4M2N1 B4JJL3 A0A1A9YDD2 A0A1B0B035 Q29HJ3 A0A195FEF0 B4L4H7 A0A1B6INM5 J9ZZN6 A0A1B0G421 A0A1B0A3H8 A0A195BG51 A0A1A9VC03 A0A195CBF6 A0A1B6DF33 A0A336LRB1 A0A1B6FII3 B4GXK7 A0A2P8XRD8 A0A1S3CXZ1 B3NV97 A0A151J979 A0A0A9XAT5 C4WX31 E0VCD4 A0A2A3ERS0 E2BID2 A0A226CXF1 A0A226E6J0 A0A0T6B6S7 A0A0N0BFP8
I4DKN9 A0A2A4JXT4 A0A2H1WLM9 A0A182H2X5 A0A023EDS6 A0A2M4AWN9 A0A2M4CJZ8 A0A2M3Z3A8 A0A2M4AWT0 A0A1S4F1U2 Q17I69 A0A2M4C2T7 B0X179 A0A1I8NDP3 A0A182WR58 A0A182LPP4 Q7Q2Z7 A0A182HNQ1 A0A0M5JDS4 A0A182VJC0 A0A182FJA9 A0A2J7PYC1 A0A067QST0 A0A1L8DP67 A0A2M4C335 A0A182MZT5 B4F5Y8 A0A182G608 B4F5Z0 Q9VZ21 B4IDZ0 B4NUA5 A0A0L0C0B6 A0A2M4AXJ8 A0A182RVP6 B4F5Z6 A0A182YH23 A0A182W5Y2 B4F5Z5 A0A182U521 B4PYE6 B4F5Z3 B3MVY1 A0A1J1ISB0 A0A0C9QQ49 A0A1W4V1L9 A0A182MST6 B4NCI9 A0A1I8PPE1 A0A2R7VU52 A0A224XPJ4 A0A161MT98 A0A182QS10 A0A084VDE1 A0A182K5P4 A0A3B0JWZ6 A0A088AE79 A0A1Y1NPU0 A0A1A9X3A5 T1ICK5 T1GCM6 A0A1W4XEN6 B4K2D2 B4M2N1 B4JJL3 A0A1A9YDD2 A0A1B0B035 Q29HJ3 A0A195FEF0 B4L4H7 A0A1B6INM5 J9ZZN6 A0A1B0G421 A0A1B0A3H8 A0A195BG51 A0A1A9VC03 A0A195CBF6 A0A1B6DF33 A0A336LRB1 A0A1B6FII3 B4GXK7 A0A2P8XRD8 A0A1S3CXZ1 B3NV97 A0A151J979 A0A0A9XAT5 C4WX31 E0VCD4 A0A2A3ERS0 E2BID2 A0A226CXF1 A0A226E6J0 A0A0T6B6S7 A0A0N0BFP8
Ontologies
GO
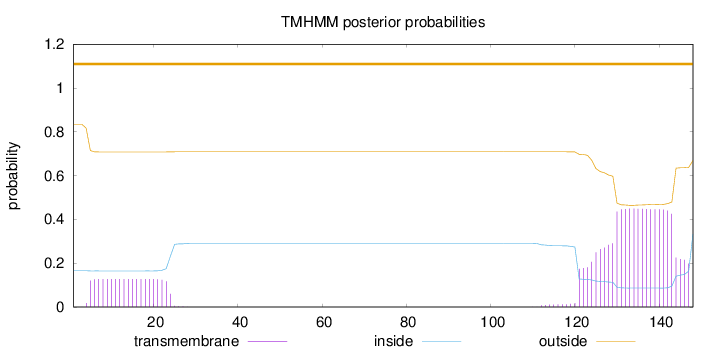
Topology
Subcellular location
Cell membrane
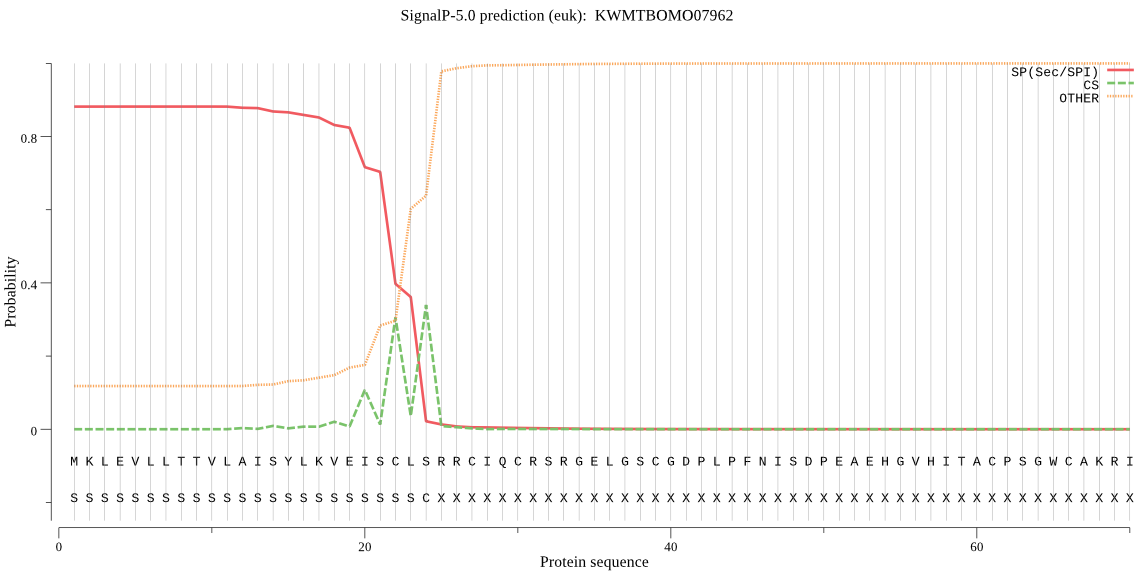
SignalP
Position: 1 - 24,
Likelihood: 0.881532
Length:
148
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.769
Exp number, first 60 AAs:
2.48665
Total prob of N-in:
0.16561
outside
1 - 148
Population Genetic Test Statistics
Pi
251.665132
Theta
221.366019
Tajima's D
0.467326
CLR
0
CSRT
0.508174591270437
Interpretation
Uncertain
