Pre Gene Modal
BGIBMGA001154
Annotation
aly_protein_[Bombyx_mori]
Full name
Protein lin-9 homolog
Alternative Name
TUDOR gene similar 1 protein
Type I interferon receptor beta chain-associated protein
Type I interferon receptor beta chain-associated protein
Location in the cell
Nuclear Reliability : 4.359
Sequence
CDS
ATGGCGGATAAATCAGAAAGAGAGTATGACGAATTATCAACTCCACCAAGATCACTCCAAAAGGTTAAAGATGAGCCAATGGAAATGGATTTTCCGGAAGAAGAAGAAGAAGAACCTCCAGCATTCGGACCTGCAGCATTGGGCTTGCATAGAGTTGGTACAAAATTGCCGCCAAAACCAGCTCCAAGTGAGCCAGTTCAAAAACTTAATGCAAGGGGAATGCCCGCTAGGATCAGAAAGAAAAATAGGTTCATTTTTGTTGACGATTTTGTGAACACGTCACCTCCACGACAGTCTCCAAAAAAGACTCCAAAAATACTTAACAAAACGCCTAATAAACCACCAAGTGCAAAAAAACAAAAATCTCCAATAAAAGTTCAAAAGGCTACAGAGAGGAATGATAAGGTAGATTCTGTAGGCATCCAAATGCAAGACAATAAATCAGGTCATCGAATTGGCATGCGACTTAGAAATTTATTGAAGCTACCCAAGGCTCACAAGTGGGTGTGTTTTGAATACTTTTATAGTAATATTGATAAAGTATTATTTGATGGTGAAAATGATTTTATGATTTGCTTAAAAGAATCATTTCCACAATTGACCAACAGAAAGTTAACTAGAACACAATGGTCAAAAATCAGAAGAATGATGGGAAAGCCTCGACGATGTTCTCAAGCATTTTTTGATGAGGAGCGGAAAGAATTGGAAAGAAAGAGGAAATTGATTCGATATGTACAACAGCAAAAATCGGCTGATGTGTGTGTCAAAGATTTACCAACCGAAATACCAATGCAATTAGTAGTTGGTACTAAAGTTACTGCTAGACTACGTCGACCACAGGATGGATTGTTCACTGGATGCATCGATGCTGTTGATACATCTAATAACACATATAGGATAACTTTTGAAAGACCTAAATTGGGCACACATTCAGTACCTGATTATGAAGTATTGTCAAATGAACCACCTGATACTATTTGTCTGACAAGTATAACGCAACGTTTTAGACCAAGAAAAGTAATTCAAGATCTACTCAGTTTGTATTCACCAATACAGAAAAATTCGCAAGGAGATCCTTTGATTGGATGTTCTGATCTTGCAAATCAAGCTAATTCTGTAATTGGTAGTTACCCATTCCGGTTTTTAGAACTCATTGTAAAATTAAGAAGATTATTAAATGCTAAAAAGAGCAAGATAAATAAATTAAAAGAATATAACTGTATGGCAGAAAAAAGAAAGTCCTTTGGACAGAGGATGCCTGAAGACTTTGAACGAAAATATGCTGCTGTTGTGATTGAGTTAGAAAGAATGAATATGGATTTACAAGAATATATCAATGAGATTCAACATCATTGTCAACAAATTGCACCAGGGCCATGTTTGGCTGCAATGTTAGCACCATCGCAACTTCGAGACAAATGTCATGAAGAAGCTTCAGTTCTTGTTGAGAAGAATAACAATGGAGTTATTCAAGATCCTTCAGTTTTAGATCTCATAACAGATCTAACTGCACTCATGTTGCAAGTTAGAAGTTTATCTGATTCAGATCAGAATGCCTATGAACTGAGTGTACTTCAAGGTACAATGGATCAGATCAAGATGAAATTAAAACCACAATATCACAGATTGTTTCAAAATAATGTTGAAATACATATGCATAAAATTCAGATGGGATTGGGGCAAATGATTTTTGAATATTCTGCAGGCACATAA
Protein
MADKSEREYDELSTPPRSLQKVKDEPMEMDFPEEEEEEPPAFGPAALGLHRVGTKLPPKPAPSEPVQKLNARGMPARIRKKNRFIFVDDFVNTSPPRQSPKKTPKILNKTPNKPPSAKKQKSPIKVQKATERNDKVDSVGIQMQDNKSGHRIGMRLRNLLKLPKAHKWVCFEYFYSNIDKVLFDGENDFMICLKESFPQLTNRKLTRTQWSKIRRMMGKPRRCSQAFFDEERKELERKRKLIRYVQQQKSADVCVKDLPTEIPMQLVVGTKVTARLRRPQDGLFTGCIDAVDTSNNTYRITFERPKLGTHSVPDYEVLSNEPPDTICLTSITQRFRPRKVIQDLLSLYSPIQKNSQGDPLIGCSDLANQANSVIGSYPFRFLELIVKLRRLLNAKKSKINKLKEYNCMAEKRKSFGQRMPEDFERKYAAVVIELERMNMDLQEYINEIQHHCQQIAPGPCLAAMLAPSQLRDKCHEEASVLVEKNNNGVIQDPSVLDLITDLTALMLQVRSLSDSDQNAYELSVLQGTMDQIKMKLKPQYHRLFQNNVEIHMHKIQMGLGQMIFEYSAGT
Summary
Description
Acts as a tumor suppressor. Inhibits DNA synthesis. Its ability to inhibit oncogenic transformation is mediated through its association with RB1. Plays a role in the expression of genes required for the G1/S transition (By similarity).
Subunit
Component of the DREAM complex (also named LINC complex) at least composed of E2F4, E2F5, LIN9, LIN37, LIN52, LIN54, MYBL1, MYBL2, RBL1, RBL2, RBBP4, TFDP1 and TFDP2. The complex exists in quiescent cells where it represses cell cycle-dependent genes. It dissociates in S phase when LIN9, LIN37, LIN52 and LIN54 form a subcomplex that binds to MYBL2. Interacts with RB1 (By similarity).
Similarity
Belongs to the lin-9 family.
Keywords
Acetylation
Alternative splicing
Cell cycle
Coiled coil
Complete proteome
DNA synthesis
Isopeptide bond
Nucleus
Phosphoprotein
Reference proteome
Tumor suppressor
Ubl conjugation
Feature
chain Protein lin-9 homolog
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
H9IV74
D0V0M8
A0A2A4JZL1
A0A1E1WKP7
A0A2H1V6N8
A0A212ETP1
+ More
S4PYF9 A0A194R4F4 A0A194QA65 A0A3S2NED6 A0A0L7LS56 A0A2J7R2K7 A0A2J7R2K0 A0A195BG63 A0A158NBV4 A0A067R349 A0A195FDE3 A0A151X9T1 E2BID1 A0A151J9K1 A0A195C9J4 A0A154P6H5 A0A1Y1LYI4 F4X8D9 E2ABC7 A0A1Y1LZH8 A0A1W4X561 A0A1Y1LTP9 A0A1W4XFT6 A0A3L8DVH9 E9J8J6 A0A0J7NSZ6 D1ZZX2 A0A2A3ETL9 K7IZB0 A0A0L7R224 A0A088AEB7 A0A0N0BFQ1 A0A232EXQ1 A0A026X131 A0A1B6FUC5 U4UDA0 N6TXS0 A0A1B6MGU0 A0A1B6D2M1 E0W435 A0A2R7VW22 A0A2R7VVH1 A0A1B0CRA5 N6UK17 T1IXW4 A0A3R7QWA5 A0A0P4WF63 A0A023F175 A0A0A9YXA9 A0A1L8DXB6 A0A1L8DWX9 A0A0A9YMA3 A0A069DVS6 Q0IFF2 A0A1S4FB31 A0A182GFR3 A0A0K2UPL5 A0A0K2UNH6 A0A182GWM1 A0A1S3K263 A0A210QWB4 B0X055 A0A226E786 V4ALJ9 K1QPE0 T1HYZ9 A0A336LMI7 C3Y680 A0A1D2MWD7 A0A146LAC4 A0A2T7PJE4 A0A3B5AY24 A0A286XYS0 A0A2D4MSX5 A0A2D4FNQ9 H0VTU6 A0A091H8T7 A0A091IE25 A0A0B6YU57 R4GCS8 A0A286XM72 K7GDQ0 A0A093IMJ2 A0A091HH70 A0A0A6YWD3 Q8C735 G1KF29 A0A093GV25 A0A2Y9JSM0 A0A3P4KT75 A0A2Y9QX57 G3SQF4 A0A0A6YVZ7
S4PYF9 A0A194R4F4 A0A194QA65 A0A3S2NED6 A0A0L7LS56 A0A2J7R2K7 A0A2J7R2K0 A0A195BG63 A0A158NBV4 A0A067R349 A0A195FDE3 A0A151X9T1 E2BID1 A0A151J9K1 A0A195C9J4 A0A154P6H5 A0A1Y1LYI4 F4X8D9 E2ABC7 A0A1Y1LZH8 A0A1W4X561 A0A1Y1LTP9 A0A1W4XFT6 A0A3L8DVH9 E9J8J6 A0A0J7NSZ6 D1ZZX2 A0A2A3ETL9 K7IZB0 A0A0L7R224 A0A088AEB7 A0A0N0BFQ1 A0A232EXQ1 A0A026X131 A0A1B6FUC5 U4UDA0 N6TXS0 A0A1B6MGU0 A0A1B6D2M1 E0W435 A0A2R7VW22 A0A2R7VVH1 A0A1B0CRA5 N6UK17 T1IXW4 A0A3R7QWA5 A0A0P4WF63 A0A023F175 A0A0A9YXA9 A0A1L8DXB6 A0A1L8DWX9 A0A0A9YMA3 A0A069DVS6 Q0IFF2 A0A1S4FB31 A0A182GFR3 A0A0K2UPL5 A0A0K2UNH6 A0A182GWM1 A0A1S3K263 A0A210QWB4 B0X055 A0A226E786 V4ALJ9 K1QPE0 T1HYZ9 A0A336LMI7 C3Y680 A0A1D2MWD7 A0A146LAC4 A0A2T7PJE4 A0A3B5AY24 A0A286XYS0 A0A2D4MSX5 A0A2D4FNQ9 H0VTU6 A0A091H8T7 A0A091IE25 A0A0B6YU57 R4GCS8 A0A286XM72 K7GDQ0 A0A093IMJ2 A0A091HH70 A0A0A6YWD3 Q8C735 G1KF29 A0A093GV25 A0A2Y9JSM0 A0A3P4KT75 A0A2Y9QX57 G3SQF4 A0A0A6YVZ7
Pubmed
19121390
22118469
23622113
26354079
26227816
21347285
+ More
24845553 20798317 28004739 21719571 30249741 21282665 18362917 19820115 20075255 28648823 24508170 23537049 20566863 25474469 25401762 26334808 17510324 26483478 28812685 23254933 22992520 18563158 27289101 26823975 21993624 17381049 19468303 21183079 16141072 16730350
24845553 20798317 28004739 21719571 30249741 21282665 18362917 19820115 20075255 28648823 24508170 23537049 20566863 25474469 25401762 26334808 17510324 26483478 28812685 23254933 22992520 18563158 27289101 26823975 21993624 17381049 19468303 21183079 16141072 16730350
EMBL
BABH01000635
GQ999610
ACY24884.1
NWSH01000379
PCG76832.1
GDQN01003623
+ More
JAT87431.1 ODYU01000707 SOQ35924.1 AGBW02012542 OWR44860.1 GAIX01003358 JAA89202.1 KQ460761 KPJ12557.1 KQ459299 KPJ01885.1 RSAL01000074 RVE48919.1 JTDY01000221 KOB78214.1 NEVH01007828 PNF35052.1 PNF35051.1 KQ976500 KYM83155.1 ADTU01011330 ADTU01011331 KK852744 KDR17349.1 KQ981673 KYN38401.1 KQ982373 KYQ57048.1 GL448504 EFN84519.1 KQ979433 KYN21569.1 KQ978068 KYM97534.1 KQ434827 KZC07462.1 GEZM01046567 JAV77075.1 GL888932 EGI57154.1 GL438237 EFN69248.1 GEZM01046566 JAV77076.1 GEZM01046568 JAV77074.1 QOIP01000004 RLU23758.1 GL769034 EFZ10847.1 LBMM01001917 KMQ95545.1 KQ971338 EFA02434.1 KZ288192 PBC34391.1 KQ414666 KOC64889.1 KQ435794 KOX73662.1 NNAY01001719 OXU23133.1 KK107036 EZA62010.1 GECZ01016020 JAS53749.1 KB632308 ERL91939.1 APGK01009034 APGK01029131 KB740725 KB737836 ENN79342.1 ENN82898.1 GEBQ01004876 JAT35101.1 GEDC01017483 JAS19815.1 DS235886 EEB20391.1 KK854082 PTY10890.1 PTY10891.1 AJWK01024544 APGK01030256 KB740764 ENN79027.1 JH431663 QCYY01001102 ROT80589.1 GDRN01061470 JAI65204.1 GBBI01003968 JAC14744.1 GBHO01039528 GBHO01009414 JAG04076.1 JAG34190.1 GFDF01003189 JAV10895.1 GFDF01003190 JAV10894.1 GBHO01039526 GBHO01009432 JAG04078.1 JAG34172.1 GBGD01001087 JAC87802.1 CH477351 EAT42873.1 JXUM01062663 KQ562210 KXJ76404.1 HACA01022451 CDW39812.1 HACA01022452 CDW39813.1 JXUM01093639 KQ564070 KXJ72837.1 NEDP02001544 OWF53011.1 DS232229 EDS37930.1 LNIX01000006 OXA52456.1 KB199650 ESP05059.1 JH818449 EKC30705.1 ACPB03022219 UFQS01000062 UFQT01000062 SSW98810.1 SSX19196.1 GG666487 EEN64477.1 LJIJ01000459 ODM97251.1 GDHC01014562 JAQ04067.1 PZQS01000003 PVD33549.1 AAKN02053936 IACM01117717 IACM01117720 LAB35863.1 IACJ01084300 LAA49115.1 KL448315 KFO82657.1 KL218535 KFP06512.1 HACG01012130 CEK58995.1 AGCU01051922 KL216060 KFV67921.1 KL537238 KFO94809.1 AC167020 AK042357 AK052618 AF190325 KL205761 KFV74158.1 CYRY02000988 VCW54932.1
JAT87431.1 ODYU01000707 SOQ35924.1 AGBW02012542 OWR44860.1 GAIX01003358 JAA89202.1 KQ460761 KPJ12557.1 KQ459299 KPJ01885.1 RSAL01000074 RVE48919.1 JTDY01000221 KOB78214.1 NEVH01007828 PNF35052.1 PNF35051.1 KQ976500 KYM83155.1 ADTU01011330 ADTU01011331 KK852744 KDR17349.1 KQ981673 KYN38401.1 KQ982373 KYQ57048.1 GL448504 EFN84519.1 KQ979433 KYN21569.1 KQ978068 KYM97534.1 KQ434827 KZC07462.1 GEZM01046567 JAV77075.1 GL888932 EGI57154.1 GL438237 EFN69248.1 GEZM01046566 JAV77076.1 GEZM01046568 JAV77074.1 QOIP01000004 RLU23758.1 GL769034 EFZ10847.1 LBMM01001917 KMQ95545.1 KQ971338 EFA02434.1 KZ288192 PBC34391.1 KQ414666 KOC64889.1 KQ435794 KOX73662.1 NNAY01001719 OXU23133.1 KK107036 EZA62010.1 GECZ01016020 JAS53749.1 KB632308 ERL91939.1 APGK01009034 APGK01029131 KB740725 KB737836 ENN79342.1 ENN82898.1 GEBQ01004876 JAT35101.1 GEDC01017483 JAS19815.1 DS235886 EEB20391.1 KK854082 PTY10890.1 PTY10891.1 AJWK01024544 APGK01030256 KB740764 ENN79027.1 JH431663 QCYY01001102 ROT80589.1 GDRN01061470 JAI65204.1 GBBI01003968 JAC14744.1 GBHO01039528 GBHO01009414 JAG04076.1 JAG34190.1 GFDF01003189 JAV10895.1 GFDF01003190 JAV10894.1 GBHO01039526 GBHO01009432 JAG04078.1 JAG34172.1 GBGD01001087 JAC87802.1 CH477351 EAT42873.1 JXUM01062663 KQ562210 KXJ76404.1 HACA01022451 CDW39812.1 HACA01022452 CDW39813.1 JXUM01093639 KQ564070 KXJ72837.1 NEDP02001544 OWF53011.1 DS232229 EDS37930.1 LNIX01000006 OXA52456.1 KB199650 ESP05059.1 JH818449 EKC30705.1 ACPB03022219 UFQS01000062 UFQT01000062 SSW98810.1 SSX19196.1 GG666487 EEN64477.1 LJIJ01000459 ODM97251.1 GDHC01014562 JAQ04067.1 PZQS01000003 PVD33549.1 AAKN02053936 IACM01117717 IACM01117720 LAB35863.1 IACJ01084300 LAA49115.1 KL448315 KFO82657.1 KL218535 KFP06512.1 HACG01012130 CEK58995.1 AGCU01051922 KL216060 KFV67921.1 KL537238 KFO94809.1 AC167020 AK042357 AK052618 AF190325 KL205761 KFV74158.1 CYRY02000988 VCW54932.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000037510 UP000235965 UP000078540 UP000005205 UP000027135 UP000078541 UP000075809 UP000008237 UP000078492 UP000078542 UP000076502 UP000007755 UP000000311 UP000192223 UP000279307 UP000036403 UP000007266 UP000242457 UP000002358 UP000053825 UP000005203 UP000053105 UP000215335 UP000053097 UP000030742 UP000019118 UP000009046 UP000092461 UP000283509 UP000008820 UP000069940 UP000249989 UP000085678 UP000242188 UP000002320 UP000198287 UP000030746 UP000005408 UP000015103 UP000001554 UP000094527 UP000245119 UP000261400 UP000005447 UP000053760 UP000054308 UP000001646 UP000007267 UP000053875 UP000000589 UP000053584 UP000248482 UP000248480 UP000007646
UP000037510 UP000235965 UP000078540 UP000005205 UP000027135 UP000078541 UP000075809 UP000008237 UP000078492 UP000078542 UP000076502 UP000007755 UP000000311 UP000192223 UP000279307 UP000036403 UP000007266 UP000242457 UP000002358 UP000053825 UP000005203 UP000053105 UP000215335 UP000053097 UP000030742 UP000019118 UP000009046 UP000092461 UP000283509 UP000008820 UP000069940 UP000249989 UP000085678 UP000242188 UP000002320 UP000198287 UP000030746 UP000005408 UP000015103 UP000001554 UP000094527 UP000245119 UP000261400 UP000005447 UP000053760 UP000054308 UP000001646 UP000007267 UP000053875 UP000000589 UP000053584 UP000248482 UP000248480 UP000007646
PRIDE
Pfam
PF06584 DIRP
ProteinModelPortal
H9IV74
D0V0M8
A0A2A4JZL1
A0A1E1WKP7
A0A2H1V6N8
A0A212ETP1
+ More
S4PYF9 A0A194R4F4 A0A194QA65 A0A3S2NED6 A0A0L7LS56 A0A2J7R2K7 A0A2J7R2K0 A0A195BG63 A0A158NBV4 A0A067R349 A0A195FDE3 A0A151X9T1 E2BID1 A0A151J9K1 A0A195C9J4 A0A154P6H5 A0A1Y1LYI4 F4X8D9 E2ABC7 A0A1Y1LZH8 A0A1W4X561 A0A1Y1LTP9 A0A1W4XFT6 A0A3L8DVH9 E9J8J6 A0A0J7NSZ6 D1ZZX2 A0A2A3ETL9 K7IZB0 A0A0L7R224 A0A088AEB7 A0A0N0BFQ1 A0A232EXQ1 A0A026X131 A0A1B6FUC5 U4UDA0 N6TXS0 A0A1B6MGU0 A0A1B6D2M1 E0W435 A0A2R7VW22 A0A2R7VVH1 A0A1B0CRA5 N6UK17 T1IXW4 A0A3R7QWA5 A0A0P4WF63 A0A023F175 A0A0A9YXA9 A0A1L8DXB6 A0A1L8DWX9 A0A0A9YMA3 A0A069DVS6 Q0IFF2 A0A1S4FB31 A0A182GFR3 A0A0K2UPL5 A0A0K2UNH6 A0A182GWM1 A0A1S3K263 A0A210QWB4 B0X055 A0A226E786 V4ALJ9 K1QPE0 T1HYZ9 A0A336LMI7 C3Y680 A0A1D2MWD7 A0A146LAC4 A0A2T7PJE4 A0A3B5AY24 A0A286XYS0 A0A2D4MSX5 A0A2D4FNQ9 H0VTU6 A0A091H8T7 A0A091IE25 A0A0B6YU57 R4GCS8 A0A286XM72 K7GDQ0 A0A093IMJ2 A0A091HH70 A0A0A6YWD3 Q8C735 G1KF29 A0A093GV25 A0A2Y9JSM0 A0A3P4KT75 A0A2Y9QX57 G3SQF4 A0A0A6YVZ7
S4PYF9 A0A194R4F4 A0A194QA65 A0A3S2NED6 A0A0L7LS56 A0A2J7R2K7 A0A2J7R2K0 A0A195BG63 A0A158NBV4 A0A067R349 A0A195FDE3 A0A151X9T1 E2BID1 A0A151J9K1 A0A195C9J4 A0A154P6H5 A0A1Y1LYI4 F4X8D9 E2ABC7 A0A1Y1LZH8 A0A1W4X561 A0A1Y1LTP9 A0A1W4XFT6 A0A3L8DVH9 E9J8J6 A0A0J7NSZ6 D1ZZX2 A0A2A3ETL9 K7IZB0 A0A0L7R224 A0A088AEB7 A0A0N0BFQ1 A0A232EXQ1 A0A026X131 A0A1B6FUC5 U4UDA0 N6TXS0 A0A1B6MGU0 A0A1B6D2M1 E0W435 A0A2R7VW22 A0A2R7VVH1 A0A1B0CRA5 N6UK17 T1IXW4 A0A3R7QWA5 A0A0P4WF63 A0A023F175 A0A0A9YXA9 A0A1L8DXB6 A0A1L8DWX9 A0A0A9YMA3 A0A069DVS6 Q0IFF2 A0A1S4FB31 A0A182GFR3 A0A0K2UPL5 A0A0K2UNH6 A0A182GWM1 A0A1S3K263 A0A210QWB4 B0X055 A0A226E786 V4ALJ9 K1QPE0 T1HYZ9 A0A336LMI7 C3Y680 A0A1D2MWD7 A0A146LAC4 A0A2T7PJE4 A0A3B5AY24 A0A286XYS0 A0A2D4MSX5 A0A2D4FNQ9 H0VTU6 A0A091H8T7 A0A091IE25 A0A0B6YU57 R4GCS8 A0A286XM72 K7GDQ0 A0A093IMJ2 A0A091HH70 A0A0A6YWD3 Q8C735 G1KF29 A0A093GV25 A0A2Y9JSM0 A0A3P4KT75 A0A2Y9QX57 G3SQF4 A0A0A6YVZ7
PDB
6C48
E-value=1.87821e-16,
Score=211
Ontologies
GO
PANTHER
Topology
Subcellular location
Found in perinucleolar structures. Associated with chromatin. With evidence from 2 publications.
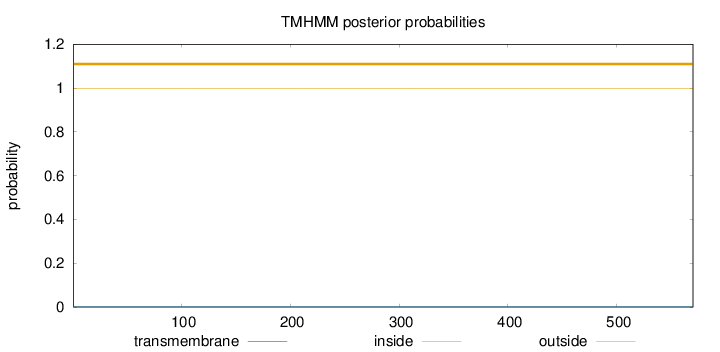
Length:
570
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000620000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00307
outside
1 - 570
Population Genetic Test Statistics
Pi
5.159964
Theta
5.430406
Tajima's D
-0.138441
CLR
0.763971
CSRT
0.351932403379831
Interpretation
Uncertain
