Gene
KWMTBOMO07960 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000995
Annotation
PREDICTED:_nuclear_pore_complex_protein_Nup50_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.41
Sequence
CDS
ATGTCTGTAAAACGGCAAGCCACGACGGATTTGAACCATGAAAATTGGGATCAAGAAGATCCTGAAGAAAATAAGGAGATGGGAACTTTTAAAGCTGCACCCAAGGATATACTGGAAAAGCGTGTTATTAAAACAGCAAAAAGACGAACGACGGCCCCTACTGATGAGAATAAAAAGACTCAAGGGCTATTCAGTGGTTTTGGTGCATTTAATAAGGCTCCATCTTCATTTGATTTTCTGGCAAATTTGACTAAAGATAGTAAAAGTAATGCTACTACAACAAGCGCTGAATCCGGATCTGCAGGATTGTTTTCAAACAAATTGTCGAATGACTCTGGTAGCCTATTGAGTCCCTCTTCTGCACCAATTAACACACCCTTAGGAACATCAACATCTCAAACACCTACTCTATTCAGTAGTGCTACTAGCACAGTTACATTTTCTTCTGTGTTTACAACTACCAAAGCAGGTCCTTCAGCTACTGATTCTCTTTTCAAAGTACAACCTACTTTGAGTACAGCCACTACTGCAGTTCTAAATGCCACAAAAACTACAACAATACCAGTTCCAAATATACAGGCTACATCTAGTATTTTTGGTGTATCACAAAACAATACAGTCAGCAAATCCTTGTTTTCAACGACAAATAACAGTTCCTCAATTTTCCAAACACAATCTACACAAAGTCCCTTAAATGATGCTACCTTTGGAACAAATAAACAATTGAATACAAACATTACTCAAGGAACTGAAAAAAAAGAGGAACAAGGTGACAAAAAATTAAAATATTATGCTAAATTGAAAGGCTTGAATGAATCTGTATCAGATTGGATTAAAAAACATGTGGATGAAACACCTTTATGCATCTTAACACCTATATTTAGAGACTATGAAGCATATTTAAAAGAAATACAGGAAGAGTATCATGGACCAGATGGTGATTGCTCTGAAATTGAGACAAAAAATAATGTTAATTCAGAAACAAAAAATCCTGTTGAAAAATCAAAAAATGGCATTTTTTCTGAATCGAGCAGAGATCAAACAGTGTCAGGATCATCAATATTTAAATTAGATCCATCTAAACCATTTTCGACCACACCATTAAGCAGTTCTTGCAATATGCTTGGAGACAAACCAAAAGGATTTTCTTTTGGTATAAATACAGCTACAAGCACTATAAATAGCATTCCAGCAATAAGCACAGGGTTCTCTTTTGGGACTTCGTCCACAAATTCCAATAATATACAAACATCAAATACCTTTTCTTTTGGATTAAAGTCTAGTCCGTCTTCTGATGGCTCTCCTCTCAAATTTGGAGCCCCATCATTAACTTCTACCAATACAAATAATACAGAGCCCGCAAATCAGGAAGATGAAGATGTACCTCCAAAAGTAGAATTTACACCAATTGCTGAAGAAAACAGTGTTTTTGACAAGCGCTGTAAAGTATTTGTGAAGAAAGATGGTAATTTTGTGGATAAAGGTGTTGGAACACTTTACATAAAAAAAATCGAAAATAGTGACAAACATCAACTACTAGTCAGAGCAAATACTGCTTTAGGTAATATACTACTTAATTTGAGGTTGTCTGCTGGTATACCTACACAAAGAATGGGTAAAAATAATGTTATGATAATATGCATTCCTACACCTGACTCTAAGCCTCCTCCCACCTCTGTATTAATAAGAGTCAAGACTCCAGAAGAAGCAGATGAAGTCCTTGAAACTTTAAACAAATATAAAGCATAA
Protein
MSVKRQATTDLNHENWDQEDPEENKEMGTFKAAPKDILEKRVIKTAKRRTTAPTDENKKTQGLFSGFGAFNKAPSSFDFLANLTKDSKSNATTTSAESGSAGLFSNKLSNDSGSLLSPSSAPINTPLGTSTSQTPTLFSSATSTVTFSSVFTTTKAGPSATDSLFKVQPTLSTATTAVLNATKTTTIPVPNIQATSSIFGVSQNNTVSKSLFSTTNNSSSIFQTQSTQSPLNDATFGTNKQLNTNITQGTEKKEEQGDKKLKYYAKLKGLNESVSDWIKKHVDETPLCILTPIFRDYEAYLKEIQEEYHGPDGDCSEIETKNNVNSETKNPVEKSKNGIFSESSRDQTVSGSSIFKLDPSKPFSTTPLSSSCNMLGDKPKGFSFGINTATSTINSIPAISTGFSFGTSSTNSNNIQTSNTFSFGLKSSPSSDGSPLKFGAPSLTSTNTNNTEPANQEDEDVPPKVEFTPIAEENSVFDKRCKVFVKKDGNFVDKGVGTLYIKKIENSDKHQLLVRANTALGNILLNLRLSAGIPTQRMGKNNVMIICIPTPDSKPPPTSVLIRVKTPEEADEVLETLNKYKA
Summary
Uniprot
Pubmed
EMBL
Proteomes
Gene 3D
ProteinModelPortal
PDB
2EC1
E-value=1.02123e-20,
Score=248
Ontologies
KEGG
GO
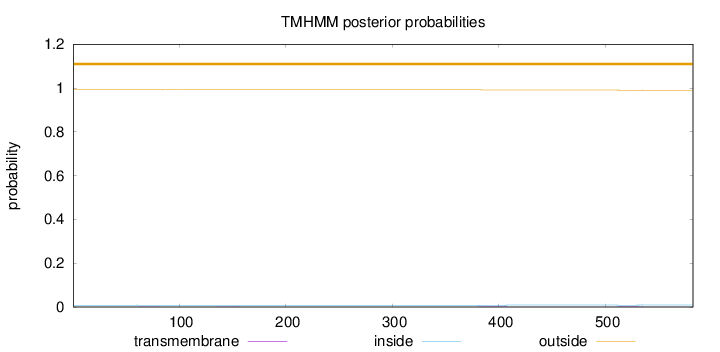
Topology
Length:
582
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09061
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.00837
outside
1 - 582
Population Genetic Test Statistics
Pi
123.389016
Theta
140.963381
Tajima's D
-0.278302
CLR
0.007456
CSRT
0.296085195740213
Interpretation
Uncertain
