Gene
KWMTBOMO07959
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.435
Sequence
CDS
ATGGACCGTGTTTGGGCGGACAGAAACATCTCGAGGAATACCAAAAAGCAGCTCGTCACCTCCCTTGTCTTTTCCATTTTTCTCTACGGCGCAGAGACTTGGACCCTCAAGAAAGCCGACCGCCAACGCATCGACGCGTTTGAGATGTGGTGCTGGAGGAAAATGCTCGGGATTCGTTGGACAGAACACAGAACCAACGAATCCATTCTTACCAAGCTCAAGATTCCAATGCGCCTCAGCACTACATGCGCGCGGAGGAGTTTCGAGATCTTTGGACATATCGCCAGAAAGAGAGGTGACAATCTGGAAAAGCTGCTGGTAACAGGCAAGGTATGCGGGAGAAGACATAGAGGCAGAAATCCAATGCGCTGGTCGGACCAAATACGCTCCACTCTCAATACCTCATTCCACGTTGCTATCCACACAGCCGAGGACAGGCAGGAATGGTGCAAGATGATGCAGGAGAAAGCTATTAGAGGAGGCCACGACCCTCAGCACTGA
Protein
MDRVWADRNISRNTKKQLVTSLVFSIFLYGAETWTLKKADRQRIDAFEMWCWRKMLGIRWTEHRTNESILTKLKIPMRLSTTCARRSFEIFGHIARKRGDNLEKLLVTGKVCGRRHRGRNPMRWSDQIRSTLNTSFHVAIHTAEDRQEWCKMMQEKAIRGGHDPQH
Summary
Uniprot
A0A2H1VNK5
A0A2H1V742
A0A2H1VU13
D7F163
D7F172
D7F161
+ More
A0A3S2L0R7 D7F168 D7F162 D7F167 D7F175 D5LB39 A0A2W1BGD9 D7F174 A0A2G8LJK7 A0A2G8K796 A0A2G8JE23 H2YWZ6 A0A2G8JVA1 H2YWZ5 A0A2G8L6P4 A0A2G8JVI4 A0A2G8K183 A0A3S3NR17 A0A3S3RLW1 A0A2G8K327 A0A3Q1MCX3 A0A2G8KTJ4 A0A3Q1LT03 W5NMM5 X1D4L4 A0A085LZ21 A0A3Q1MZC9 A0A3S3P648 A0A3Q1MGA3 A0A3Q1LFQ0 A0A3Q1LLH3 W4Y921 A0A3Q1M4E7 W5NIE2 A0A3Q1LR75 O97916 W5MM77 A0A3Q1NE26 A0A3Q1MJR3 F6R5D0 A0A3Q1M7G0 W4YPU3 A0A3Q1MJK8 A0A3Q1M4J9 A0A3Q1M2T8 A0A3Q1MFU8 A0A3Q1MJZ2 A0A3Q1MXT5 A0A3Q1MI63 A0A3Q1MNS1 A0A3Q1MIT2 A0A3Q1MQ02 A0A3Q1MN71 A0A3Q1LQN1 A0A3Q1MMS2 A0A3Q1LUA6 A0A3Q1MHE1 A0A3Q1LNR9 A0A3Q1MDY8 J9LZK0 A0A3Q1NB85 X1WK45 A0A3S1HJZ0 A0A3Q1LRK8 A0A3Q1N9K5 A0A3Q1N8A1 A0A2H6KKM9 A0A3Q1LNZ2 A0A3Q1MMW6 A0A3Q1M6Y4 A0A3Q1N3Y9 W5MXL2 A0A023G885 A0A085NA37 A0A3Q1NFA5 A0A3Q1NE48 W4XFH1 A0A3Q1MLV4 A0A3Q1LWU3 A0A2W1BSS7 A0A3Q1ML90 A0A3Q1LKL4 A0A3Q1MVY4 A0A3Q1LKN4 J9LIJ7 A0A3Q1LGI2 A0A3Q1LT63 A0A067RQM5 A0A3Q1MN00 A0A2S2QAF2
A0A3S2L0R7 D7F168 D7F162 D7F167 D7F175 D5LB39 A0A2W1BGD9 D7F174 A0A2G8LJK7 A0A2G8K796 A0A2G8JE23 H2YWZ6 A0A2G8JVA1 H2YWZ5 A0A2G8L6P4 A0A2G8JVI4 A0A2G8K183 A0A3S3NR17 A0A3S3RLW1 A0A2G8K327 A0A3Q1MCX3 A0A2G8KTJ4 A0A3Q1LT03 W5NMM5 X1D4L4 A0A085LZ21 A0A3Q1MZC9 A0A3S3P648 A0A3Q1MGA3 A0A3Q1LFQ0 A0A3Q1LLH3 W4Y921 A0A3Q1M4E7 W5NIE2 A0A3Q1LR75 O97916 W5MM77 A0A3Q1NE26 A0A3Q1MJR3 F6R5D0 A0A3Q1M7G0 W4YPU3 A0A3Q1MJK8 A0A3Q1M4J9 A0A3Q1M2T8 A0A3Q1MFU8 A0A3Q1MJZ2 A0A3Q1MXT5 A0A3Q1MI63 A0A3Q1MNS1 A0A3Q1MIT2 A0A3Q1MQ02 A0A3Q1MN71 A0A3Q1LQN1 A0A3Q1MMS2 A0A3Q1LUA6 A0A3Q1MHE1 A0A3Q1LNR9 A0A3Q1MDY8 J9LZK0 A0A3Q1NB85 X1WK45 A0A3S1HJZ0 A0A3Q1LRK8 A0A3Q1N9K5 A0A3Q1N8A1 A0A2H6KKM9 A0A3Q1LNZ2 A0A3Q1MMW6 A0A3Q1M6Y4 A0A3Q1N3Y9 W5MXL2 A0A023G885 A0A085NA37 A0A3Q1NFA5 A0A3Q1NE48 W4XFH1 A0A3Q1MLV4 A0A3Q1LWU3 A0A2W1BSS7 A0A3Q1ML90 A0A3Q1LKL4 A0A3Q1MVY4 A0A3Q1LKN4 J9LIJ7 A0A3Q1LGI2 A0A3Q1LT63 A0A067RQM5 A0A3Q1MN00 A0A2S2QAF2
EMBL
ODYU01003523
SOQ42419.1
ODYU01001007
SOQ36619.1
ODYU01004401
SOQ44236.1
+ More
FJ265548 ADI61816.1 FJ265557 ADI61825.1 FJ265546 ADI61814.1 RSAL01000289 RVE42931.1 FJ265553 ADI61821.1 FJ265547 ADI61815.1 FJ265552 ADI61820.1 FJ265560 ADI61828.1 GU815090 ADF18553.1 KZ150203 PZC72267.1 FJ265559 ADI61827.1 MRZV01000056 PIK60437.1 MRZV01000817 PIK43880.1 MRZV01002322 PIK33998.1 MRZV01001216 PIK39650.1 MRZV01000196 PIK55924.1 MRZV01001207 PIK39730.1 MRZV01000994 PIK41715.1 NCKU01012182 RWS00163.1 NCKU01008836 RWS01655.1 MRZV01000940 PIK42365.1 MRZV01000378 PIK51329.1 AHAT01022938 BART01031562 GAH15691.1 KL363256 KL367985 KFD50217.1 KFD59415.1 NCKU01004395 RWS06003.1 AAGJ04161766 AHAT01009145 AJ132772 CAA10770.1 AHAT01015286 AAGJ04055194 ABLF02042206 ABLF02024992 ABLF02024994 ABLF02025002 ABLF02043975 RQTK01000360 RUS81011.1 BDSA01000081 GBE63537.1 AHAT01017596 GBBM01006298 JAC29120.1 KL363266 KL367526 KFD49675.1 KFD66333.1 AAGJ04075245 KZ149985 PZC75696.1 ABLF02012409 ABLF02012412 KK852482 KDR22935.1 GGMS01004979 MBY74182.1
FJ265548 ADI61816.1 FJ265557 ADI61825.1 FJ265546 ADI61814.1 RSAL01000289 RVE42931.1 FJ265553 ADI61821.1 FJ265547 ADI61815.1 FJ265552 ADI61820.1 FJ265560 ADI61828.1 GU815090 ADF18553.1 KZ150203 PZC72267.1 FJ265559 ADI61827.1 MRZV01000056 PIK60437.1 MRZV01000817 PIK43880.1 MRZV01002322 PIK33998.1 MRZV01001216 PIK39650.1 MRZV01000196 PIK55924.1 MRZV01001207 PIK39730.1 MRZV01000994 PIK41715.1 NCKU01012182 RWS00163.1 NCKU01008836 RWS01655.1 MRZV01000940 PIK42365.1 MRZV01000378 PIK51329.1 AHAT01022938 BART01031562 GAH15691.1 KL363256 KL367985 KFD50217.1 KFD59415.1 NCKU01004395 RWS06003.1 AAGJ04161766 AHAT01009145 AJ132772 CAA10770.1 AHAT01015286 AAGJ04055194 ABLF02042206 ABLF02024992 ABLF02024994 ABLF02025002 ABLF02043975 RQTK01000360 RUS81011.1 BDSA01000081 GBE63537.1 AHAT01017596 GBBM01006298 JAC29120.1 KL363266 KL367526 KFD49675.1 KFD66333.1 AAGJ04075245 KZ149985 PZC75696.1 ABLF02012409 ABLF02012412 KK852482 KDR22935.1 GGMS01004979 MBY74182.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2H1VNK5
A0A2H1V742
A0A2H1VU13
D7F163
D7F172
D7F161
+ More
A0A3S2L0R7 D7F168 D7F162 D7F167 D7F175 D5LB39 A0A2W1BGD9 D7F174 A0A2G8LJK7 A0A2G8K796 A0A2G8JE23 H2YWZ6 A0A2G8JVA1 H2YWZ5 A0A2G8L6P4 A0A2G8JVI4 A0A2G8K183 A0A3S3NR17 A0A3S3RLW1 A0A2G8K327 A0A3Q1MCX3 A0A2G8KTJ4 A0A3Q1LT03 W5NMM5 X1D4L4 A0A085LZ21 A0A3Q1MZC9 A0A3S3P648 A0A3Q1MGA3 A0A3Q1LFQ0 A0A3Q1LLH3 W4Y921 A0A3Q1M4E7 W5NIE2 A0A3Q1LR75 O97916 W5MM77 A0A3Q1NE26 A0A3Q1MJR3 F6R5D0 A0A3Q1M7G0 W4YPU3 A0A3Q1MJK8 A0A3Q1M4J9 A0A3Q1M2T8 A0A3Q1MFU8 A0A3Q1MJZ2 A0A3Q1MXT5 A0A3Q1MI63 A0A3Q1MNS1 A0A3Q1MIT2 A0A3Q1MQ02 A0A3Q1MN71 A0A3Q1LQN1 A0A3Q1MMS2 A0A3Q1LUA6 A0A3Q1MHE1 A0A3Q1LNR9 A0A3Q1MDY8 J9LZK0 A0A3Q1NB85 X1WK45 A0A3S1HJZ0 A0A3Q1LRK8 A0A3Q1N9K5 A0A3Q1N8A1 A0A2H6KKM9 A0A3Q1LNZ2 A0A3Q1MMW6 A0A3Q1M6Y4 A0A3Q1N3Y9 W5MXL2 A0A023G885 A0A085NA37 A0A3Q1NFA5 A0A3Q1NE48 W4XFH1 A0A3Q1MLV4 A0A3Q1LWU3 A0A2W1BSS7 A0A3Q1ML90 A0A3Q1LKL4 A0A3Q1MVY4 A0A3Q1LKN4 J9LIJ7 A0A3Q1LGI2 A0A3Q1LT63 A0A067RQM5 A0A3Q1MN00 A0A2S2QAF2
A0A3S2L0R7 D7F168 D7F162 D7F167 D7F175 D5LB39 A0A2W1BGD9 D7F174 A0A2G8LJK7 A0A2G8K796 A0A2G8JE23 H2YWZ6 A0A2G8JVA1 H2YWZ5 A0A2G8L6P4 A0A2G8JVI4 A0A2G8K183 A0A3S3NR17 A0A3S3RLW1 A0A2G8K327 A0A3Q1MCX3 A0A2G8KTJ4 A0A3Q1LT03 W5NMM5 X1D4L4 A0A085LZ21 A0A3Q1MZC9 A0A3S3P648 A0A3Q1MGA3 A0A3Q1LFQ0 A0A3Q1LLH3 W4Y921 A0A3Q1M4E7 W5NIE2 A0A3Q1LR75 O97916 W5MM77 A0A3Q1NE26 A0A3Q1MJR3 F6R5D0 A0A3Q1M7G0 W4YPU3 A0A3Q1MJK8 A0A3Q1M4J9 A0A3Q1M2T8 A0A3Q1MFU8 A0A3Q1MJZ2 A0A3Q1MXT5 A0A3Q1MI63 A0A3Q1MNS1 A0A3Q1MIT2 A0A3Q1MQ02 A0A3Q1MN71 A0A3Q1LQN1 A0A3Q1MMS2 A0A3Q1LUA6 A0A3Q1MHE1 A0A3Q1LNR9 A0A3Q1MDY8 J9LZK0 A0A3Q1NB85 X1WK45 A0A3S1HJZ0 A0A3Q1LRK8 A0A3Q1N9K5 A0A3Q1N8A1 A0A2H6KKM9 A0A3Q1LNZ2 A0A3Q1MMW6 A0A3Q1M6Y4 A0A3Q1N3Y9 W5MXL2 A0A023G885 A0A085NA37 A0A3Q1NFA5 A0A3Q1NE48 W4XFH1 A0A3Q1MLV4 A0A3Q1LWU3 A0A2W1BSS7 A0A3Q1ML90 A0A3Q1LKL4 A0A3Q1MVY4 A0A3Q1LKN4 J9LIJ7 A0A3Q1LGI2 A0A3Q1LT63 A0A067RQM5 A0A3Q1MN00 A0A2S2QAF2
Ontologies
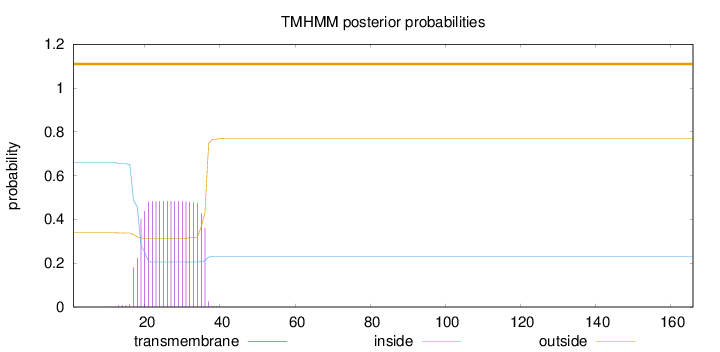
Topology
Length:
166
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.83407999999999
Exp number, first 60 AAs:
8.8339
Total prob of N-in:
0.66132
outside
1 - 166
Population Genetic Test Statistics
Pi
354.509312
Theta
154.182042
Tajima's D
3.994298
CLR
0
CSRT
0.999200039998
Interpretation
Uncertain
