Gene
KWMTBOMO07957
Pre Gene Modal
BGIBMGA000994
Annotation
PREDICTED:_malate_dehydrogenase_1?_mitochondrial-like_[Bombyx_mori]
Full name
Malate dehydrogenase
Location in the cell
Mitochondrial Reliability : 1.679 PlasmaMembrane Reliability : 1.016
Sequence
CDS
ATGATAGTGAATCGAAGAATAATCTCGTCGTTGTTCACCACAACAAGAAGTTACCAAGTTACGGTAGTTGGCGCAACAAGCGAAGTCGGTCAAGTCCTGAGTCTATTACTGAGATCTTTACCTTTAATAACTAAATTAGTTGTTCACGATAACAAGGAATTTACTCCAGGTGTTACATTAGATTTATCACAAATACCCAGTCCCTCGCGAATATTAGGATTGACTGGCGAAGATACCTTAGAACGAGCTCTTAAAAACTCTGATTTGATCATAGCAGCTGGCGGACTTTCACAACGTCCAGATATAAGTGAAAAAGCTCTGTTGACAGCGAATAGCCAGTTCATTAAGTCTATCATCTCGAAATTGGGAAGATCGTGTCCTATACCTTTTGTAGGAATAGTTACTGAACCTATAAACTATTTAATTCCCATGACTGCTGAAATTATGAGGAATCACGGTGTTTATGATGAAAAAAAGTTATTTGGAATAACCGAAATTGACGCCATACGATCGCAGTGTCTTTACGCCACGGAGAATAATTTGAAAGCAAACGATTGTTATGTACCTGTTATTGGAGGCCATTCAGATAAAACGATTGTCCCATTGTTGTCTCAAGCTAAACCCACTGTGGAAATGAAAGAAAAAGATTTAGAAGAATTTACCATTAAATTGCGAAATTGCGATGGTATGATAACAAAGGCTAAAAGAGGGTGTATTCCTAATTTGTCAGTTGCATATAGTAGTTTTCTTTTCACTAAAAGCATTCTCGACGCTTTAGAAGGCAGCCCTGCAAAAATTCACGCTTTTGTAGATAACAATGACTTTGGTACATCATATTTCTCTAGCCTGGTGAATCTGGATAAAAATGGCGTCAAGGAAATGGTAAGATATTCAGAGTTCTCAAAATTTGAGTGTGATTTAATTGAAAAAAGTTTGCAGCAGTTGCGAAAAGACGTATCTAAAGGAAGGAAAATATTGGAACTGGCGTAA
Protein
MIVNRRIISSLFTTTRSYQVTVVGATSEVGQVLSLLLRSLPLITKLVVHDNKEFTPGVTLDLSQIPSPSRILGLTGEDTLERALKNSDLIIAAGGLSQRPDISEKALLTANSQFIKSIISKLGRSCPIPFVGIVTEPINYLIPMTAEIMRNHGVYDEKKLFGITEIDAIRSQCLYATENNLKANDCYVPVIGGHSDKTIVPLLSQAKPTVEMKEKDLEEFTIKLRNCDGMITKAKRGCIPNLSVAYSSFLFTKSILDALEGSPAKIHAFVDNNDFGTSYFSSLVNLDKNGVKEMVRYSEFSKFECDLIEKSLQQLRKDVSKGRKILELA
Summary
Description
Catalyzes the reversible oxidation of malate to oxaloacetate.
Catalytic Activity
(S)-malate + NAD(+) = H(+) + NADH + oxaloacetate
Subunit
Homodimer.
Similarity
Belongs to the LDH/MDH superfamily.
Belongs to the LDH/MDH superfamily. MDH type 1 family.
Belongs to the LDH/MDH superfamily. MDH type 1 family.
Feature
chain Malate dehydrogenase
Uniprot
H9IUR4
A0A0L7LSP5
A0A2A4JXU5
A0A2H1W725
A0A194R5Q1
A0A194Q8N5
+ More
A0A1I7VM37 A0A069DYP3 A0A158Q7N8 A0A0K0IZA2 A0A0N4TT58 A0A1I8CFH1 A0A0V0G3N9 A0A1L8EI00 A0A0K0EJS9 J9F386 A0A0H5SP26 A0A0N4ZV85 D8RBW3 A0A0L0BXC1 A0A151KTC4 A0A099MAH7 A0A0F2I4B9 A0A087U9C0 A0A1I8B0F3 D8RXR3 A0A0K0F735 A0A0N5B542 A0A3R9EK95 A0A3P6SGY9 A0A023FKC0 A0A1Y0D1C4 U4K8H6 A0A2N0XS07 A0A090LKA7 A0A1Y6IZK4 Q4PP90 A0A2G8L9N0 A0A3B1JQM0 A0A1L8EEQ2 A0A1L8EEK1 A0A3B0KCD1 A0A2P7R8R3 Q9NHX3 A0A0L7ZFB9 D2A663 A0A1S3ZW53 A0A1U7YI28 A0A023FJ07 A0A0C9S2L6 R4G866 A0A329EAC4 U4DXT9 B4K7H4 A0A0C9QFA6 A0A034VDU0 A0A0K8VWB8 A0A023FWQ5 A0A317ZXA6 A0A2N6CU64 A0A3S0Q063 A0A1J6IJL7 V5SJ21 A0A170ZKM6 A0A182DZC4 R7TSV0 A0A1V0I515 B4NG95 A0A026WP45 A0A380NUH3 C9P2S3 A0A2P0XJ03 A0A2K6WIX3 A0A2G5CGV8 A0A2J8GIH2 W8BVV6 A0A1W4U941 D0X9A1 A0A086WHW1 A0A097QN77 B4IB85 Q9VEB1 A0A1S4A9C7 A0A2I4ETB2 D1MBR2 B4PL86 A0A3L6FDC0 B4FRJ1 A0A2Z5RE49 A0A231MX69 K3XJN7 A0A2G9UBR2 Q6BCF2 Q6BCF0 A0A0P7XGH5 A0A3S4Q176 A0A3P8VIR3 A0A1B0AIQ6 B4M0W4
A0A1I7VM37 A0A069DYP3 A0A158Q7N8 A0A0K0IZA2 A0A0N4TT58 A0A1I8CFH1 A0A0V0G3N9 A0A1L8EI00 A0A0K0EJS9 J9F386 A0A0H5SP26 A0A0N4ZV85 D8RBW3 A0A0L0BXC1 A0A151KTC4 A0A099MAH7 A0A0F2I4B9 A0A087U9C0 A0A1I8B0F3 D8RXR3 A0A0K0F735 A0A0N5B542 A0A3R9EK95 A0A3P6SGY9 A0A023FKC0 A0A1Y0D1C4 U4K8H6 A0A2N0XS07 A0A090LKA7 A0A1Y6IZK4 Q4PP90 A0A2G8L9N0 A0A3B1JQM0 A0A1L8EEQ2 A0A1L8EEK1 A0A3B0KCD1 A0A2P7R8R3 Q9NHX3 A0A0L7ZFB9 D2A663 A0A1S3ZW53 A0A1U7YI28 A0A023FJ07 A0A0C9S2L6 R4G866 A0A329EAC4 U4DXT9 B4K7H4 A0A0C9QFA6 A0A034VDU0 A0A0K8VWB8 A0A023FWQ5 A0A317ZXA6 A0A2N6CU64 A0A3S0Q063 A0A1J6IJL7 V5SJ21 A0A170ZKM6 A0A182DZC4 R7TSV0 A0A1V0I515 B4NG95 A0A026WP45 A0A380NUH3 C9P2S3 A0A2P0XJ03 A0A2K6WIX3 A0A2G5CGV8 A0A2J8GIH2 W8BVV6 A0A1W4U941 D0X9A1 A0A086WHW1 A0A097QN77 B4IB85 Q9VEB1 A0A1S4A9C7 A0A2I4ETB2 D1MBR2 B4PL86 A0A3L6FDC0 B4FRJ1 A0A2Z5RE49 A0A231MX69 K3XJN7 A0A2G9UBR2 Q6BCF2 Q6BCF0 A0A0P7XGH5 A0A3S4Q176 A0A3P8VIR3 A0A1B0AIQ6 B4M0W4
EC Number
1.1.1.37
Pubmed
19121390
26227816
26354079
25217238
26334808
17885136
+ More
26829753 26850696 21551031 26108605 18809916 23739050 27311813 29023486 25329095 10713448 18362917 19820115 24807620 23773524 26131772 17994087 25348373 22919073 23254933 24508170 24495485 19860885 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27145194 17550304 30061735 19936069 26760975 22580951 16598154 30626928 24487278
26829753 26850696 21551031 26108605 18809916 23739050 27311813 29023486 25329095 10713448 18362917 19820115 24807620 23773524 26131772 17994087 25348373 22919073 23254933 24508170 24495485 19860885 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27145194 17550304 30061735 19936069 26760975 22580951 16598154 30626928 24487278
EMBL
BABH01000643
JTDY01000221
KOB78221.1
NWSH01000379
PCG76837.1
ODYU01006624
+ More
SOQ48632.1 KQ460761 KPJ12560.1 KQ459299 KPJ01888.1 JH712099 EFO26441.1 GBGD01002040 JAC86849.1 UZAD01013255 VDN93060.1 GECL01003432 JAP02692.1 GFDG01000570 JAV18229.1 ADBV01000013 UYWW01000109 EJW88983.1 VDM07303.1 LN857006 CRZ25392.1 GL377575 EFJ30575.1 JRES01001197 KNC24655.1 LOBR01000109 KYN81875.1 JMCG01000001 KGK10254.1 JZAM01000168 KJR31831.1 KK118832 KFM73959.1 GL377594 EFJ22957.1 RSFA01000010 RSD32391.1 UYRX01000105 VDK74056.1 GBBK01002903 JAC21579.1 CP021376 ART80905.1 FO203526 CCO58964.1 PIZL01000019 PKF78653.1 LN609529 CEF67990.1 FXXI01000010 SMS02451.1 DQ062223 AAY63978.1 MRZV01000159 PIK56961.1 GFDG01001643 JAV17156.1 GFDG01001652 JAV17147.1 OUUW01000008 SPP83849.1 PXYG01000002 PSJ46605.1 AF218064 AAF27650.1 LFWL01000049 KOE88878.1 KQ971346 EFA04972.1 GBBK01002880 JAC21602.1 GBZX01001486 JAG91254.1 ACPB03000504 GAHY01001507 JAA76003.1 QLTR01000009 RAS64464.1 CANW01000002 CCN39022.1 CH933806 EDW15318.1 GBYB01013203 JAG82970.1 GAKP01018685 GAKP01018684 JAC40267.1 GDHF01009138 JAI43176.1 GBBL01001391 JAC25929.1 QHMB01000011 PXA69912.1 PKUN01000023 PLX60668.1 RXZH01000009 RTZ14337.1 MJEQ01037190 OIS99074.1 KF647639 AHB50501.1 GEMB01002082 JAS01097.1 UYRW01000119 VDK63335.1 AMQN01012226 KB309380 ELT94561.1 CP020453 ARC91281.1 CH964251 EDW83312.1 KK107139 EZA57723.1 UHIS01000006 SUP51126.1 ACZO01000006 EEX37971.1 KY661311 AVA17398.1 CMVM020000250 KZ305072 PIA30524.1 POSH01000020 PNH85793.1 GAMC01003193 JAC03363.1 ACZC01000015 EEZ88439.1 JPQB01000033 KFI09361.1 NRHY01000021 NRQO01000019 PAU35767.1 PAW01028.1 CH480827 EDW44643.1 AE014297 AY119152 BT029274 AAF55516.1 AAM51012.1 ABK30911.1 GU130143 ACZ13336.1 CM000160 EDW95868.1 NCVQ01000004 PWZ31152.1 BT039729 CM007649 ACF84734.1 ONM39824.1 FX983743 BAX07242.1 CP012621 NMUO01000056 ATG75149.1 OXS14784.1 AGNK02003222 CM003532 RCV26435.1 KZ347463 PIO67669.1 AB185122 BAD30065.1 AB185123 BAD30066.1 LIHO01000011 KPQ02561.1 QPKB01000013 RWR97473.1 CH940650 EDW68423.1
SOQ48632.1 KQ460761 KPJ12560.1 KQ459299 KPJ01888.1 JH712099 EFO26441.1 GBGD01002040 JAC86849.1 UZAD01013255 VDN93060.1 GECL01003432 JAP02692.1 GFDG01000570 JAV18229.1 ADBV01000013 UYWW01000109 EJW88983.1 VDM07303.1 LN857006 CRZ25392.1 GL377575 EFJ30575.1 JRES01001197 KNC24655.1 LOBR01000109 KYN81875.1 JMCG01000001 KGK10254.1 JZAM01000168 KJR31831.1 KK118832 KFM73959.1 GL377594 EFJ22957.1 RSFA01000010 RSD32391.1 UYRX01000105 VDK74056.1 GBBK01002903 JAC21579.1 CP021376 ART80905.1 FO203526 CCO58964.1 PIZL01000019 PKF78653.1 LN609529 CEF67990.1 FXXI01000010 SMS02451.1 DQ062223 AAY63978.1 MRZV01000159 PIK56961.1 GFDG01001643 JAV17156.1 GFDG01001652 JAV17147.1 OUUW01000008 SPP83849.1 PXYG01000002 PSJ46605.1 AF218064 AAF27650.1 LFWL01000049 KOE88878.1 KQ971346 EFA04972.1 GBBK01002880 JAC21602.1 GBZX01001486 JAG91254.1 ACPB03000504 GAHY01001507 JAA76003.1 QLTR01000009 RAS64464.1 CANW01000002 CCN39022.1 CH933806 EDW15318.1 GBYB01013203 JAG82970.1 GAKP01018685 GAKP01018684 JAC40267.1 GDHF01009138 JAI43176.1 GBBL01001391 JAC25929.1 QHMB01000011 PXA69912.1 PKUN01000023 PLX60668.1 RXZH01000009 RTZ14337.1 MJEQ01037190 OIS99074.1 KF647639 AHB50501.1 GEMB01002082 JAS01097.1 UYRW01000119 VDK63335.1 AMQN01012226 KB309380 ELT94561.1 CP020453 ARC91281.1 CH964251 EDW83312.1 KK107139 EZA57723.1 UHIS01000006 SUP51126.1 ACZO01000006 EEX37971.1 KY661311 AVA17398.1 CMVM020000250 KZ305072 PIA30524.1 POSH01000020 PNH85793.1 GAMC01003193 JAC03363.1 ACZC01000015 EEZ88439.1 JPQB01000033 KFI09361.1 NRHY01000021 NRQO01000019 PAU35767.1 PAW01028.1 CH480827 EDW44643.1 AE014297 AY119152 BT029274 AAF55516.1 AAM51012.1 ABK30911.1 GU130143 ACZ13336.1 CM000160 EDW95868.1 NCVQ01000004 PWZ31152.1 BT039729 CM007649 ACF84734.1 ONM39824.1 FX983743 BAX07242.1 CP012621 NMUO01000056 ATG75149.1 OXS14784.1 AGNK02003222 CM003532 RCV26435.1 KZ347463 PIO67669.1 AB185122 BAD30065.1 AB185123 BAD30066.1 LIHO01000011 KPQ02561.1 QPKB01000013 RWR97473.1 CH940650 EDW68423.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000095285
+ More
UP000050640 UP000006672 UP000038020 UP000278627 UP000095286 UP000035681 UP000004810 UP000093561 UP000270924 UP000038045 UP000001514 UP000037069 UP000075346 UP000029994 UP000033477 UP000054359 UP000095281 UP000035680 UP000046392 UP000269041 UP000277928 UP000243793 UP000016895 UP000233580 UP000035682 UP000196125 UP000230750 UP000018467 UP000268350 UP000240243 UP000037536 UP000007266 UP000084051 UP000189701 UP000015103 UP000248729 UP000018247 UP000009192 UP000246709 UP000235015 UP000268973 UP000187609 UP000077448 UP000271087 UP000014760 UP000007798 UP000053097 UP000254349 UP000005604 UP000024404 UP000230069 UP000236330 UP000192221 UP000004396 UP000029061 UP000217861 UP000218364 UP000001292 UP000000803 UP000235220 UP000002282 UP000251960 UP000007305 UP000215233 UP000217763 UP000004995 UP000053932 UP000283530 UP000265120 UP000092445 UP000008792
UP000050640 UP000006672 UP000038020 UP000278627 UP000095286 UP000035681 UP000004810 UP000093561 UP000270924 UP000038045 UP000001514 UP000037069 UP000075346 UP000029994 UP000033477 UP000054359 UP000095281 UP000035680 UP000046392 UP000269041 UP000277928 UP000243793 UP000016895 UP000233580 UP000035682 UP000196125 UP000230750 UP000018467 UP000268350 UP000240243 UP000037536 UP000007266 UP000084051 UP000189701 UP000015103 UP000248729 UP000018247 UP000009192 UP000246709 UP000235015 UP000268973 UP000187609 UP000077448 UP000271087 UP000014760 UP000007798 UP000053097 UP000254349 UP000005604 UP000024404 UP000230069 UP000236330 UP000192221 UP000004396 UP000029061 UP000217861 UP000218364 UP000001292 UP000000803 UP000235220 UP000002282 UP000251960 UP000007305 UP000215233 UP000217763 UP000004995 UP000053932 UP000283530 UP000265120 UP000092445 UP000008792
Interpro
Gene 3D
ProteinModelPortal
H9IUR4
A0A0L7LSP5
A0A2A4JXU5
A0A2H1W725
A0A194R5Q1
A0A194Q8N5
+ More
A0A1I7VM37 A0A069DYP3 A0A158Q7N8 A0A0K0IZA2 A0A0N4TT58 A0A1I8CFH1 A0A0V0G3N9 A0A1L8EI00 A0A0K0EJS9 J9F386 A0A0H5SP26 A0A0N4ZV85 D8RBW3 A0A0L0BXC1 A0A151KTC4 A0A099MAH7 A0A0F2I4B9 A0A087U9C0 A0A1I8B0F3 D8RXR3 A0A0K0F735 A0A0N5B542 A0A3R9EK95 A0A3P6SGY9 A0A023FKC0 A0A1Y0D1C4 U4K8H6 A0A2N0XS07 A0A090LKA7 A0A1Y6IZK4 Q4PP90 A0A2G8L9N0 A0A3B1JQM0 A0A1L8EEQ2 A0A1L8EEK1 A0A3B0KCD1 A0A2P7R8R3 Q9NHX3 A0A0L7ZFB9 D2A663 A0A1S3ZW53 A0A1U7YI28 A0A023FJ07 A0A0C9S2L6 R4G866 A0A329EAC4 U4DXT9 B4K7H4 A0A0C9QFA6 A0A034VDU0 A0A0K8VWB8 A0A023FWQ5 A0A317ZXA6 A0A2N6CU64 A0A3S0Q063 A0A1J6IJL7 V5SJ21 A0A170ZKM6 A0A182DZC4 R7TSV0 A0A1V0I515 B4NG95 A0A026WP45 A0A380NUH3 C9P2S3 A0A2P0XJ03 A0A2K6WIX3 A0A2G5CGV8 A0A2J8GIH2 W8BVV6 A0A1W4U941 D0X9A1 A0A086WHW1 A0A097QN77 B4IB85 Q9VEB1 A0A1S4A9C7 A0A2I4ETB2 D1MBR2 B4PL86 A0A3L6FDC0 B4FRJ1 A0A2Z5RE49 A0A231MX69 K3XJN7 A0A2G9UBR2 Q6BCF2 Q6BCF0 A0A0P7XGH5 A0A3S4Q176 A0A3P8VIR3 A0A1B0AIQ6 B4M0W4
A0A1I7VM37 A0A069DYP3 A0A158Q7N8 A0A0K0IZA2 A0A0N4TT58 A0A1I8CFH1 A0A0V0G3N9 A0A1L8EI00 A0A0K0EJS9 J9F386 A0A0H5SP26 A0A0N4ZV85 D8RBW3 A0A0L0BXC1 A0A151KTC4 A0A099MAH7 A0A0F2I4B9 A0A087U9C0 A0A1I8B0F3 D8RXR3 A0A0K0F735 A0A0N5B542 A0A3R9EK95 A0A3P6SGY9 A0A023FKC0 A0A1Y0D1C4 U4K8H6 A0A2N0XS07 A0A090LKA7 A0A1Y6IZK4 Q4PP90 A0A2G8L9N0 A0A3B1JQM0 A0A1L8EEQ2 A0A1L8EEK1 A0A3B0KCD1 A0A2P7R8R3 Q9NHX3 A0A0L7ZFB9 D2A663 A0A1S3ZW53 A0A1U7YI28 A0A023FJ07 A0A0C9S2L6 R4G866 A0A329EAC4 U4DXT9 B4K7H4 A0A0C9QFA6 A0A034VDU0 A0A0K8VWB8 A0A023FWQ5 A0A317ZXA6 A0A2N6CU64 A0A3S0Q063 A0A1J6IJL7 V5SJ21 A0A170ZKM6 A0A182DZC4 R7TSV0 A0A1V0I515 B4NG95 A0A026WP45 A0A380NUH3 C9P2S3 A0A2P0XJ03 A0A2K6WIX3 A0A2G5CGV8 A0A2J8GIH2 W8BVV6 A0A1W4U941 D0X9A1 A0A086WHW1 A0A097QN77 B4IB85 Q9VEB1 A0A1S4A9C7 A0A2I4ETB2 D1MBR2 B4PL86 A0A3L6FDC0 B4FRJ1 A0A2Z5RE49 A0A231MX69 K3XJN7 A0A2G9UBR2 Q6BCF2 Q6BCF0 A0A0P7XGH5 A0A3S4Q176 A0A3P8VIR3 A0A1B0AIQ6 B4M0W4
PDB
4E0B
E-value=3.97633e-46,
Score=465
Ontologies
PATHWAY
00020
Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
PANTHER
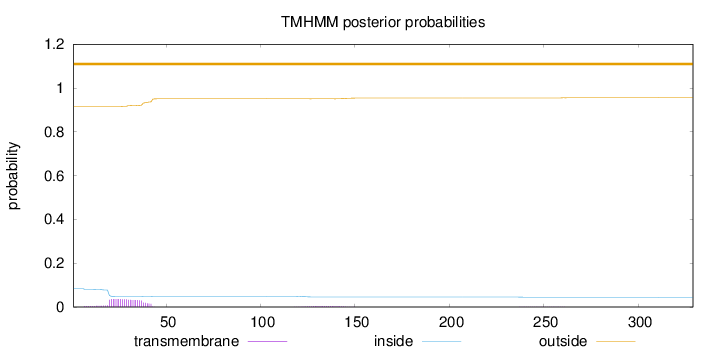
Topology
Length:
329
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.864849999999999
Exp number, first 60 AAs:
0.75494
Total prob of N-in:
0.08407
outside
1 - 329
Population Genetic Test Statistics
Pi
52.409584
Theta
124.699184
Tajima's D
-0.460984
CLR
5.878747
CSRT
0.247287635618219
Interpretation
Uncertain
