Gene
KWMTBOMO07950
Pre Gene Modal
BGIBMGA000990
Annotation
EXD1_BOMMO_RecName:_Full%3DpiRNA_biogenesis_protein_EXD1;_short=BmExd1;_altname:_full=Exonuclease_3'-5'_domain-containing_protein_1,Exonuclease_3'-5'_domain-like-containing_protein_1,Inactive_exonuclease_EXD1
Full name
piRNA biogenesis protein EXD1
Alternative Name
Exonuclease 3'-5' domain-containing protein 1
Exonuclease 3'-5' domain-like-containing protein 1
Inactive exonuclease EXD1
Exonuclease 3'-5' domain-like-containing protein 1
Inactive exonuclease EXD1
Location in the cell
Cytoplasmic Reliability : 2.869
Sequence
CDS
ATGGACAACCTCTACACAAAAGGAGAACTGTTACAAGTTCACACAAAAAACTACGACGTCTTTGAAGGTAGATTCTATAGTATGGCCCAAGATAAGACCAAAATATCACTATATGATGTCAAGGAAATTCCACATGGTGATGCTAATGATGGAGTCCTTCATTACTATGATTCTGAAATAAGAGAAGTGGTGAAACTCCAGGAGTCTACTGAAAAGAAAGTTCTCAAAATATCGCAAACGAAATATGAAGAAATCCTCAAAATATCAAAAAAATACATTTTCATAAACCAAGTTGATAAAAGTTTCCATGAAGCTGTGGATGATTTGAACCAGCAAGATTTTATTGCTGTTAGCGGTGATGGTGCAAATATGGGGAGAAAATGCAAGATGCCATTTCTTGTTTTGTCCACTGACCACCAGATCTATATCTTTGACATCCAGGTGATGCAATATCATGCATTTGAATCAGGCTTAAAAAAAATTTTGGAAGGTGATTCTCCAAAAAAGATTGCACATGATTGTAGAAAATTGTCTGATTGTCTATATCATAAACACAATGTTAAGCTGAAATCAGTTTTTGATACTCAGGTTGGTGATTTAATAATCACTAAGAATAAGAAGGTCACTTTACCAAACAAAGTGAGATCTCTGGGAGAATGTTTGACCAATTACCTGGGTCTACAACAAAACACCATTGACGAAAAGCTTGATATTGTTCAAAGTACAGAAAGGCCACTTTCAGTGAAGATCAAGGATAGTCTGGCAAGGAATATTGCATTTTTACACCATCTGTCTGAAGTTATTAATGAAGAAATGCAGTTACCGTTTTACAGAGGTGTTGAATGCTATATTGAAAACATTCGTTCTTCAGACGATTTTAAAGCCTGGGAACTATGTGGAAAATTAAATCAAATACCAAAGGAGTTTAGAAACGCTATTGACTATTAA
Protein
MDNLYTKGELLQVHTKNYDVFEGRFYSMAQDKTKISLYDVKEIPHGDANDGVLHYYDSEIREVVKLQESTEKKVLKISQTKYEEILKISKKYIFINQVDKSFHEAVDDLNQQDFIAVSGDGANMGRKCKMPFLVLSTDHQIYIFDIQVMQYHAFESGLKKILEGDSPKKIAHDCRKLSDCLYHKHNVKLKSVFDTQVGDLIITKNKKVTLPNKVRSLGECLTNYLGLQQNTIDEKLDIVQSTERPLSVKIKDSLARNIAFLHHLSEVINEEMQLPFYRGVECYIENIRSSDDFKAWELCGKLNQIPKEFRNAIDY
Summary
Description
RNA-binding component of the PET complex, a multiprotein complex required for the processing of piRNAs during spermatogenesis. The piRNA metabolic process mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins and governs the methylation and subsequent repression of transposable elements, preventing their mobilization, which is essential for the germline integrity. The PET complex is required during the secondary piRNAs metabolic process for the PIWIL2 slicing-triggered loading of PIWIL4 piRNAs. In the PET complex, EXD1 probably acts as an RNA adapter. EXD1 is an inactive exonuclease.
RNA-binding component of the PET complex, a multiprotein complex required for the processing of piRNAs during spermatogenesis. The piRNA metabolic process mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins and governs the methylation and subsequent repression of transposable elements, preventing their mobilization, which is essential for the germline integrity (By similarity). The PET complex is required during the secondary piRNAs metabolic process for the PIWIL2 slicing-triggered loading of PIWIL4 piRNAs. In the PET complex, EXD1 probably acts as an RNA adapter. EXD1 is an inactive exonuclease (By similarity).
RNA-binding component of the PET complex, a multiprotein complex required for the processing of piRNAs during spermatogenesis. The piRNA metabolic process mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins and governs the methylation and subsequent repression of transposable elements, preventing their mobilization, which is essential for the germline integrity (By similarity). The PET complex is required during the secondary piRNAs metabolic process for the PIWIL2 slicing-triggered loading of PIWIL4 piRNAs. In the PET complex, EXD1 probably acts as an RNA adapter. EXD1 is an inactive exonuclease (By similarity).
Subunit
Homodimer (PubMed:26669262). Component of the PET complex, at least composed of EXD1, SIWI, TDRD12 and piRNAs (PubMed:26669262).
Homodimer (By similarity). Component of the PET complex, at least composed of EXD1, PIWIL2, TDRD12 and piRNAs (By similarity).
Homodimer (By similarity). Component of the PET complex, at least composed of EXD1, PIWIL2, TDRD12 and piRNAs (By similarity).
Similarity
Belongs to the EXD1 family.
Keywords
3D-structure
Complete proteome
Cytoplasm
Meiosis
Reference proteome
RNA-binding
RNA-mediated gene silencing
Alternative splicing
Polymorphism
Feature
chain piRNA biogenesis protein EXD1
splice variant In isoform 2.
sequence variant In dbSNP:rs522063.
splice variant In isoform 2.
sequence variant In dbSNP:rs522063.
Uniprot
H9IUR0
A0A158N7N9
A0A212FA25
S4NNP1
A0A2A4JVM0
A0A194R435
+ More
A0A2H1W061 A0A194QEK3 A0A0L7L1U9 A0A2J7RFV2 A0A2J7RFV8 A0A1W4XPM8 A0A067QUM6 A0A067R1Q8 A0A2J7PN93 B7P8W9 V5H9Q5 A0A131YAC7 K1QXC0 A0A224YSX7 A0A2R5LME7 A0A131YVU9 A0A2T7PLX6 N6TAR9 U4UN96 A0A2G8LI45 T1IXR4 R7VH01 A0A293N4R6 A0A151MUZ2 A0A2B4SEQ1 A0A151MV39 A0A1E1XBB6 G1KC49 A0A3M6UWT9 A0A182WCL0 G1NF98 A0A182LZ97 A0A2D4M251 I3NEZ6 A0A2D4M239 A0A2D4L3L2 D2GW91 G1MEF5 A0A2P8Y281 A0A0T6BAJ1 A0A3Q0GSE7 A0A182RXG7 A0A182YI45 A0A3Q0GMW3 A0A1U7RJR5 A0A1U8D3K9 F6XAA5 A0A1U8D3F1 A0A2K5YQW3 W4YND1 A0A1S3AKJ8 A0A087V1P4 A0A2K5YQW0 A0A2K6LTF7 G3WW68 H0VH70 Q16PQ9 A0A1S3HIB3 A0A1S3HF89 G1QTB8 A0A383ZZ90 A0A2K6LTE4 A0A2Y9HHZ2 A0A1S4FU23 A0A2I3H0S1 A0A384D131 S7MHY0 A0A3Q7TIB9 D4AD21 G1T664 A0A2Y9IRE6 A0A2U3XZ50 A0A384D081 A0A3Q7UI02 A0A287APJ8 A0A3Q7QBM5 Q8NHP7 G1NVN8 H2NMW3 A0A2I2ZA85 L5K1S9 A0A2K6CW17
A0A2H1W061 A0A194QEK3 A0A0L7L1U9 A0A2J7RFV2 A0A2J7RFV8 A0A1W4XPM8 A0A067QUM6 A0A067R1Q8 A0A2J7PN93 B7P8W9 V5H9Q5 A0A131YAC7 K1QXC0 A0A224YSX7 A0A2R5LME7 A0A131YVU9 A0A2T7PLX6 N6TAR9 U4UN96 A0A2G8LI45 T1IXR4 R7VH01 A0A293N4R6 A0A151MUZ2 A0A2B4SEQ1 A0A151MV39 A0A1E1XBB6 G1KC49 A0A3M6UWT9 A0A182WCL0 G1NF98 A0A182LZ97 A0A2D4M251 I3NEZ6 A0A2D4M239 A0A2D4L3L2 D2GW91 G1MEF5 A0A2P8Y281 A0A0T6BAJ1 A0A3Q0GSE7 A0A182RXG7 A0A182YI45 A0A3Q0GMW3 A0A1U7RJR5 A0A1U8D3K9 F6XAA5 A0A1U8D3F1 A0A2K5YQW3 W4YND1 A0A1S3AKJ8 A0A087V1P4 A0A2K5YQW0 A0A2K6LTF7 G3WW68 H0VH70 Q16PQ9 A0A1S3HIB3 A0A1S3HF89 G1QTB8 A0A383ZZ90 A0A2K6LTE4 A0A2Y9HHZ2 A0A1S4FU23 A0A2I3H0S1 A0A384D131 S7MHY0 A0A3Q7TIB9 D4AD21 G1T664 A0A2Y9IRE6 A0A2U3XZ50 A0A384D081 A0A3Q7UI02 A0A287APJ8 A0A3Q7QBM5 Q8NHP7 G1NVN8 H2NMW3 A0A2I2ZA85 L5K1S9 A0A2K6CW17
Pubmed
19121390
26669262
22118469
23622113
26354079
26227816
+ More
24845553 25765539 22992520 28797301 26830274 23537049 29023486 23254933 22293439 28503490 30382153 20838655 20010809 29403074 25244985 17495919 21709235 21993624 17510324 15057822 15632090 24813606 14702039 16572171 15489334 22398555 23258410
24845553 25765539 22992520 28797301 26830274 23537049 29023486 23254933 22293439 28503490 30382153 20838655 20010809 29403074 25244985 17495919 21709235 21993624 17510324 15057822 15632090 24813606 14702039 16572171 15489334 22398555 23258410
EMBL
BABH01000658
AGBW02009529
OWR50580.1
GAIX01013851
JAA78709.1
NWSH01000547
+ More
PCG75738.1 KQ460761 KPJ12568.1 ODYU01005447 SOQ46316.1 KQ459299 KPJ01896.1 JTDY01003524 KOB69410.1 NEVH01004410 PNF39707.1 PNF39708.1 KK852917 KDR13862.1 KK852811 KDR15932.1 NEVH01023957 PNF17798.1 ABJB010207539 ABJB010295509 ABJB010501444 DS660824 EEC03041.1 GANP01004438 JAB80030.1 GEFM01000396 JAP75400.1 JH818278 EKC38348.1 GFPF01007703 MAA18849.1 GGLE01006574 MBY10700.1 GEDV01005885 JAP82672.1 PZQS01000003 PVD34404.1 APGK01037606 KB740948 ENN77374.1 KB632288 ERL91480.1 MRZV01000070 PIK59924.1 JH431661 AMQN01004538 KB293746 ELU15581.1 GFWV01023316 MAA48043.1 AKHW03004924 KYO28371.1 LSMT01000112 PFX27037.1 KYO28372.1 GFAC01002648 JAT96540.1 RCHS01000557 RMX57984.1 AXCM01001478 IACM01070106 IACM01070108 LAB27410.1 AGTP01002270 AGTP01002271 AGTP01002272 AGTP01002273 AGTP01002274 AGTP01002275 IACM01070111 IACM01070112 LAB27424.1 IACL01113034 LAB15433.1 GL192356 EFB13799.1 ACTA01040374 PYGN01001027 PSN38340.1 LJIG01002654 KRT84286.1 AAGJ04111396 AAGJ04111397 KL867280 KFM83533.1 AEFK01010259 AEFK01010260 AEFK01010261 AAKN02015898 AAKN02015899 CH477774 EAT36364.1 ADFV01034090 ADFV01034091 ADFV01034092 ADFV01034093 ADFV01034094 ADFV01034095 ADFV01034096 ADFV01034097 ADFV01034098 ADFV01034099 KE161462 EPQ03984.1 AABR07072914 CH473949 EDL79907.1 AAGW02024529 AEMK02000004 AK123414 AK292524 AK302864 AC012652 AC022408 BC030628 AAPE02025717 ABGA01338794 ABGA01338795 ABGA01338796 ABGA01338797 ABGA01338798 ABGA01338799 ABGA01338800 ABGA01338801 ABGA01338802 ABGA01338803 NDHI03003606 PNJ16164.1 CABD030095600 CABD030095601 CABD030095602 KB031042 ELK05525.1
PCG75738.1 KQ460761 KPJ12568.1 ODYU01005447 SOQ46316.1 KQ459299 KPJ01896.1 JTDY01003524 KOB69410.1 NEVH01004410 PNF39707.1 PNF39708.1 KK852917 KDR13862.1 KK852811 KDR15932.1 NEVH01023957 PNF17798.1 ABJB010207539 ABJB010295509 ABJB010501444 DS660824 EEC03041.1 GANP01004438 JAB80030.1 GEFM01000396 JAP75400.1 JH818278 EKC38348.1 GFPF01007703 MAA18849.1 GGLE01006574 MBY10700.1 GEDV01005885 JAP82672.1 PZQS01000003 PVD34404.1 APGK01037606 KB740948 ENN77374.1 KB632288 ERL91480.1 MRZV01000070 PIK59924.1 JH431661 AMQN01004538 KB293746 ELU15581.1 GFWV01023316 MAA48043.1 AKHW03004924 KYO28371.1 LSMT01000112 PFX27037.1 KYO28372.1 GFAC01002648 JAT96540.1 RCHS01000557 RMX57984.1 AXCM01001478 IACM01070106 IACM01070108 LAB27410.1 AGTP01002270 AGTP01002271 AGTP01002272 AGTP01002273 AGTP01002274 AGTP01002275 IACM01070111 IACM01070112 LAB27424.1 IACL01113034 LAB15433.1 GL192356 EFB13799.1 ACTA01040374 PYGN01001027 PSN38340.1 LJIG01002654 KRT84286.1 AAGJ04111396 AAGJ04111397 KL867280 KFM83533.1 AEFK01010259 AEFK01010260 AEFK01010261 AAKN02015898 AAKN02015899 CH477774 EAT36364.1 ADFV01034090 ADFV01034091 ADFV01034092 ADFV01034093 ADFV01034094 ADFV01034095 ADFV01034096 ADFV01034097 ADFV01034098 ADFV01034099 KE161462 EPQ03984.1 AABR07072914 CH473949 EDL79907.1 AAGW02024529 AEMK02000004 AK123414 AK292524 AK302864 AC012652 AC022408 BC030628 AAPE02025717 ABGA01338794 ABGA01338795 ABGA01338796 ABGA01338797 ABGA01338798 ABGA01338799 ABGA01338800 ABGA01338801 ABGA01338802 ABGA01338803 NDHI03003606 PNJ16164.1 CABD030095600 CABD030095601 CABD030095602 KB031042 ELK05525.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000053268
UP000037510
+ More
UP000235965 UP000192223 UP000027135 UP000001555 UP000005408 UP000245119 UP000019118 UP000030742 UP000230750 UP000014760 UP000050525 UP000225706 UP000001646 UP000275408 UP000075920 UP000001645 UP000075883 UP000005215 UP000008912 UP000245037 UP000189705 UP000075900 UP000076408 UP000002280 UP000233140 UP000007110 UP000079721 UP000054359 UP000233180 UP000007648 UP000005447 UP000008820 UP000085678 UP000001073 UP000248481 UP000261680 UP000286642 UP000002494 UP000001811 UP000248482 UP000245341 UP000291021 UP000008227 UP000286641 UP000005640 UP000001074 UP000001595 UP000001519 UP000010552 UP000233120
UP000235965 UP000192223 UP000027135 UP000001555 UP000005408 UP000245119 UP000019118 UP000030742 UP000230750 UP000014760 UP000050525 UP000225706 UP000001646 UP000275408 UP000075920 UP000001645 UP000075883 UP000005215 UP000008912 UP000245037 UP000189705 UP000075900 UP000076408 UP000002280 UP000233140 UP000007110 UP000079721 UP000054359 UP000233180 UP000007648 UP000005447 UP000008820 UP000085678 UP000001073 UP000248481 UP000261680 UP000286642 UP000002494 UP000001811 UP000248482 UP000245341 UP000291021 UP000008227 UP000286641 UP000005640 UP000001074 UP000001595 UP000001519 UP000010552 UP000233120
Pfam
Interpro
IPR036397
RNaseH_sf
+ More
IPR012337 RNaseH-like_sf
IPR002562 3'-5'_exonuclease_dom
IPR000421 FA58C
IPR009030 Growth_fac_rcpt_cys_sf
IPR016186 C-type_lectin-like/link_sf
IPR013320 ConA-like_dom_sf
IPR036055 LDL_receptor-like_sf
IPR035914 Sperma_CUB_dom_sf
IPR002172 LDrepeatLR_classA_rpt
IPR000859 CUB_dom
IPR001304 C-type_lectin-like
IPR001881 EGF-like_Ca-bd_dom
IPR035976 Sushi/SCR/CCP_sf
IPR011641 Tyr-kin_ephrin_A/B_rcpt-like
IPR023415 LDLR_class-A_CS
IPR003410 HYR_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR000742 EGF-like_dom
IPR000436 Sushi_SCR_CCP_dom
IPR016187 CTDL_fold
IPR008979 Galactose-bd-like_sf
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR025609 Lsm14-like_N
IPR012337 RNaseH-like_sf
IPR002562 3'-5'_exonuclease_dom
IPR000421 FA58C
IPR009030 Growth_fac_rcpt_cys_sf
IPR016186 C-type_lectin-like/link_sf
IPR013320 ConA-like_dom_sf
IPR036055 LDL_receptor-like_sf
IPR035914 Sperma_CUB_dom_sf
IPR002172 LDrepeatLR_classA_rpt
IPR000859 CUB_dom
IPR001304 C-type_lectin-like
IPR001881 EGF-like_Ca-bd_dom
IPR035976 Sushi/SCR/CCP_sf
IPR011641 Tyr-kin_ephrin_A/B_rcpt-like
IPR023415 LDLR_class-A_CS
IPR003410 HYR_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR000742 EGF-like_dom
IPR000436 Sushi_SCR_CCP_dom
IPR016187 CTDL_fold
IPR008979 Galactose-bd-like_sf
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR025609 Lsm14-like_N
SUPFAM
ProteinModelPortal
H9IUR0
A0A158N7N9
A0A212FA25
S4NNP1
A0A2A4JVM0
A0A194R435
+ More
A0A2H1W061 A0A194QEK3 A0A0L7L1U9 A0A2J7RFV2 A0A2J7RFV8 A0A1W4XPM8 A0A067QUM6 A0A067R1Q8 A0A2J7PN93 B7P8W9 V5H9Q5 A0A131YAC7 K1QXC0 A0A224YSX7 A0A2R5LME7 A0A131YVU9 A0A2T7PLX6 N6TAR9 U4UN96 A0A2G8LI45 T1IXR4 R7VH01 A0A293N4R6 A0A151MUZ2 A0A2B4SEQ1 A0A151MV39 A0A1E1XBB6 G1KC49 A0A3M6UWT9 A0A182WCL0 G1NF98 A0A182LZ97 A0A2D4M251 I3NEZ6 A0A2D4M239 A0A2D4L3L2 D2GW91 G1MEF5 A0A2P8Y281 A0A0T6BAJ1 A0A3Q0GSE7 A0A182RXG7 A0A182YI45 A0A3Q0GMW3 A0A1U7RJR5 A0A1U8D3K9 F6XAA5 A0A1U8D3F1 A0A2K5YQW3 W4YND1 A0A1S3AKJ8 A0A087V1P4 A0A2K5YQW0 A0A2K6LTF7 G3WW68 H0VH70 Q16PQ9 A0A1S3HIB3 A0A1S3HF89 G1QTB8 A0A383ZZ90 A0A2K6LTE4 A0A2Y9HHZ2 A0A1S4FU23 A0A2I3H0S1 A0A384D131 S7MHY0 A0A3Q7TIB9 D4AD21 G1T664 A0A2Y9IRE6 A0A2U3XZ50 A0A384D081 A0A3Q7UI02 A0A287APJ8 A0A3Q7QBM5 Q8NHP7 G1NVN8 H2NMW3 A0A2I2ZA85 L5K1S9 A0A2K6CW17
A0A2H1W061 A0A194QEK3 A0A0L7L1U9 A0A2J7RFV2 A0A2J7RFV8 A0A1W4XPM8 A0A067QUM6 A0A067R1Q8 A0A2J7PN93 B7P8W9 V5H9Q5 A0A131YAC7 K1QXC0 A0A224YSX7 A0A2R5LME7 A0A131YVU9 A0A2T7PLX6 N6TAR9 U4UN96 A0A2G8LI45 T1IXR4 R7VH01 A0A293N4R6 A0A151MUZ2 A0A2B4SEQ1 A0A151MV39 A0A1E1XBB6 G1KC49 A0A3M6UWT9 A0A182WCL0 G1NF98 A0A182LZ97 A0A2D4M251 I3NEZ6 A0A2D4M239 A0A2D4L3L2 D2GW91 G1MEF5 A0A2P8Y281 A0A0T6BAJ1 A0A3Q0GSE7 A0A182RXG7 A0A182YI45 A0A3Q0GMW3 A0A1U7RJR5 A0A1U8D3K9 F6XAA5 A0A1U8D3F1 A0A2K5YQW3 W4YND1 A0A1S3AKJ8 A0A087V1P4 A0A2K5YQW0 A0A2K6LTF7 G3WW68 H0VH70 Q16PQ9 A0A1S3HIB3 A0A1S3HF89 G1QTB8 A0A383ZZ90 A0A2K6LTE4 A0A2Y9HHZ2 A0A1S4FU23 A0A2I3H0S1 A0A384D131 S7MHY0 A0A3Q7TIB9 D4AD21 G1T664 A0A2Y9IRE6 A0A2U3XZ50 A0A384D081 A0A3Q7UI02 A0A287APJ8 A0A3Q7QBM5 Q8NHP7 G1NVN8 H2NMW3 A0A2I2ZA85 L5K1S9 A0A2K6CW17
PDB
5FIS
E-value=5.62245e-133,
Score=1214
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Component of the meiotic nuage, also named P granule, a germ-cell-specific organelle required to repress transposon activity during meiosis. With evidence from 4 publications.
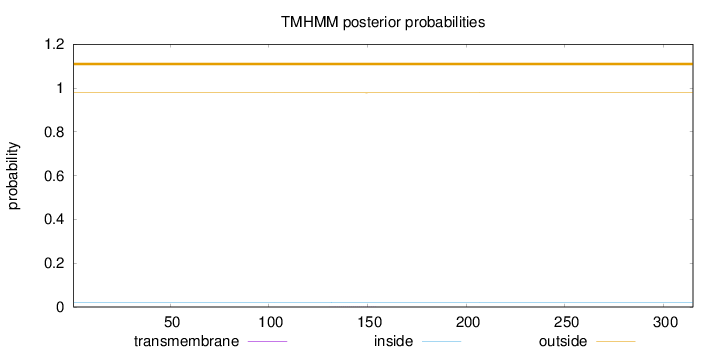
Length:
315
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01412
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02191
outside
1 - 315
Population Genetic Test Statistics
Pi
177.597696
Theta
162.408883
Tajima's D
0.279644
CLR
0
CSRT
0.449577521123944
Interpretation
Uncertain
