Gene
KWMTBOMO07948
Pre Gene Modal
BGIBMGA001160
Annotation
putative_glutamine-dependent_NAD(+)_synthetase_[Papilio_machaon]
Full name
Glutamine-dependent NAD(+) synthetase
Alternative Name
NAD(+) synthase [glutamine-hydrolyzing]
Location in the cell
Nuclear Reliability : 1.485 PlasmaMembrane Reliability : 2.031
Sequence
CDS
ATGGGTAGAAAGGTGACAGTAGCGGTTTGTACGTTGAATCAGTGGGCTCTGGACTTTGAAGGAAATTTGAACAGGATATTACAGTCAATCCAAGAAGCCAAAGAGCTCGGTGCACTGTATCGAACTGGCCCGGAGTTAGAGATATGTGGTTATAGCTGCGAAGATCACTTCCACGAATCCGACACGTACCTTCACAGCTGGCAAGTGCTAGTCGAACTATTGAAATCGCCAACATGTAAAGACATACTCATAGACGTCGGTATGCCGGTACAGCACCGTAACGTCTCATACAATTGTCGTGTGGCGTTTTTCAACAGGAAGATAATACTTATCAGACCGAAAATGATATTGTGTGACGACGGGAATTACAGGGAGACCAGATGGTTCTCCTGTTGGACTAAAGACAGACAAGTCGAAGACTTTTATTTACCAAGAATGATAACGGCAGTGACGAACCAATCCACCGTTCCTATTGGTGACGCGGTTATCTCGACCAGAGACACTTGTATCGGTTTCGAAATATGCGAAGAATTGTGGAATCCACAAAGCCGCCACATTCCGTTGAGCTTGGACGGGGTAGAAATAATATCGAATGGCTCCGGCAGCTATATGGAGCTCCGTAAAGCGTACGTCACGGTCGACTTGGTGAAGAGCGCGACCTTCAAGAGCGGGGGCGCATATTTGTTCAGCAACTTGAGGGGCTGCGACGGCCAGAGGATATACTTCAACGGCTGCTCGTGTGTGGCCGTGAACGGAGAAATTGTCAGTCGGGGGCAGCAGTTCGGCTTGATCGATGTTGAAGTAACGACCGCTACCATAGATTTAGAAGATATAAGGTCTTATCGAGCAAAAAATAGATCTCGTTGTCATTTGGCAGCTTCGAATAAACCATTTCCCAGGATCTTTGTGGATGTTTCGCTGTCTGACGATGAAGACATACATTTAACGACGAATCCACCTATACAATGGCACTATTTGAGTCCAGAAGAGGAAATATCTTTGGGTCCCGCGTGCTGGCTGTGGGACTACCTCCGTCGCTCGGGCCAGGGCGGCTTCTTTCTACCGTTGAGCGGCGGAGTGGACTCCAGCAGCACCGCATGCATTGTGTTCTCTATGTGCACACAGATATGCGAAGCCATTAAGAAAGGAGAAAGTCAAGTGTTATATGATGTAAGGAAAATCATGTGTCAGCCTGACTACACACCGTCCGACCCGATGGAGCTGTGCAACAAGCTACTGGTTACTTGTTATATGGCGTCGGAGAACTCGAGCCGCGAGACCAAGCAGAGAGCTTCGCAGCTCGCAAGCCAGATAGGCAGTTACCATTTCCCAATTCTGATCGATACGGCGGTTAATGCTGCGCTTGGGATCTTTACTGCGGCCACTGGTCTTTTACCGATATTCAAAAGCAAAGGAGGCTGCCCGAGACAGAACCTTGCATTACAAAATATACAAGTAAGCATCGAAAACAATAAATAA
Protein
MGRKVTVAVCTLNQWALDFEGNLNRILQSIQEAKELGALYRTGPELEICGYSCEDHFHESDTYLHSWQVLVELLKSPTCKDILIDVGMPVQHRNVSYNCRVAFFNRKIILIRPKMILCDDGNYRETRWFSCWTKDRQVEDFYLPRMITAVTNQSTVPIGDAVISTRDTCIGFEICEELWNPQSRHIPLSLDGVEIISNGSGSYMELRKAYVTVDLVKSATFKSGGAYLFSNLRGCDGQRIYFNGCSCVAVNGEIVSRGQQFGLIDVEVTTATIDLEDIRSYRAKNRSRCHLAASNKPFPRIFVDVSLSDDEDIHLTTNPPIQWHYLSPEEEISLGPACWLWDYLRRSGQGGFFLPLSGGVDSSSTACIVFSMCTQICEAIKKGESQVLYDVRKIMCQPDYTPSDPMELCNKLLVTCYMASENSSRETKQRASQLASQIGSYHFPILIDTAVNAALGIFTAATGLLPIFKSKGGCPRQNLALQNIQVSIENNK
Summary
Description
Catalyzes the ATP-dependent amidation of deamido-NAD to form NAD. Uses L-glutamine as a nitrogen source (By similarity). Because of its role in energy metabolism, involved in the modulation of aged-related cardiac function, mobility, and lifespan (PubMed:27448710).
Catalytic Activity
ATP + deamido-NAD(+) + H2O + L-glutamine = AMP + diphosphate + H(+) + L-glutamate + NAD(+)
Similarity
In the C-terminal section; belongs to the NAD synthetase family.
Keywords
ATP-binding
Complete proteome
Ligase
NAD
Nucleotide-binding
Phosphoprotein
Reference proteome
Feature
chain Glutamine-dependent NAD(+) synthetase
Uniprot
H9IV80
A0A194R9P4
A0A194Q906
A0A3S2M6H4
A0A2H1WA98
A0A2H1VEG0
+ More
A0A2A4JVD5 A0A212EY75 A0A1Q3G1P3 A0A1Q3G1Q7 A0A336KV63 T1PKA9 A0A1J1HV96 D1ZZT1 A0A1I8NS65 A0A2J7RRY1 B4JL37 A0A1W4XJK8 Q29HW0 B4GY71 A0A0M3QZQ0 A0A067R837 A0A3B0JXK5 A0A034VB33 B4NC89 B4R4E5 A0A0K8W6T7 B4Q2E8 B4IG53 C5WLN1 Q9VYA0 A0A1W4VGV4 K7IZ08 A0A0A1XD61 A0A232F2S6 B3MW01 B4L7J5 A0A0Q9WMA9 B4M2T6 B3NWK7 A0A195BW65 A0A088AIB9 A0A158NCC5 A0A151I6Z7 A0A0L7RDJ9 A0A2A3ELI5 F4WX79 A0A151J5W2 E9J843 E2B1W6 A0A026WUW9 A0A1B0G4F1 A0A195F9M6 A0A1B0AHA3 A0A1A9WFA1 A0A1A9VWF6 A0A1A9YF37 A0A1B0C7C8 A0A154P6M5 E0VPD9 A0A1Y1KLN7 A0A310SFQ0 A0A1B6KSA4 A0A1B6JRC4 A0A151WFS0 A0A182QC98 A0A182MPM7 A0A182YQW1 E2BGL8 A0A182W416 A0A182RVT2 A0A084WFJ9 A0A182KGG7 A0A0P4WDD5 A0A182N052 A0A182JFP2 L7M2U8 U4U8I8 A0A182HIJ6 A0A182U6G3 A0A182WY73 V5HYE3 A0A087TML8 A0A182VMG5 A0A1S3JZK0 A0A2R5L8C7 Q7PS02 Q16Z66 A0A0M9A797 A0A1L8GIT4 A0A0L0C3M6 K1Q3V6 A0A0B6ZQ45 A0A023F4V6 A0A0N8A8E5 F7DFI1 B1H2T4 G3TWX8 A0A0N8DV92 A0A0P6H6W2
A0A2A4JVD5 A0A212EY75 A0A1Q3G1P3 A0A1Q3G1Q7 A0A336KV63 T1PKA9 A0A1J1HV96 D1ZZT1 A0A1I8NS65 A0A2J7RRY1 B4JL37 A0A1W4XJK8 Q29HW0 B4GY71 A0A0M3QZQ0 A0A067R837 A0A3B0JXK5 A0A034VB33 B4NC89 B4R4E5 A0A0K8W6T7 B4Q2E8 B4IG53 C5WLN1 Q9VYA0 A0A1W4VGV4 K7IZ08 A0A0A1XD61 A0A232F2S6 B3MW01 B4L7J5 A0A0Q9WMA9 B4M2T6 B3NWK7 A0A195BW65 A0A088AIB9 A0A158NCC5 A0A151I6Z7 A0A0L7RDJ9 A0A2A3ELI5 F4WX79 A0A151J5W2 E9J843 E2B1W6 A0A026WUW9 A0A1B0G4F1 A0A195F9M6 A0A1B0AHA3 A0A1A9WFA1 A0A1A9VWF6 A0A1A9YF37 A0A1B0C7C8 A0A154P6M5 E0VPD9 A0A1Y1KLN7 A0A310SFQ0 A0A1B6KSA4 A0A1B6JRC4 A0A151WFS0 A0A182QC98 A0A182MPM7 A0A182YQW1 E2BGL8 A0A182W416 A0A182RVT2 A0A084WFJ9 A0A182KGG7 A0A0P4WDD5 A0A182N052 A0A182JFP2 L7M2U8 U4U8I8 A0A182HIJ6 A0A182U6G3 A0A182WY73 V5HYE3 A0A087TML8 A0A182VMG5 A0A1S3JZK0 A0A2R5L8C7 Q7PS02 Q16Z66 A0A0M9A797 A0A1L8GIT4 A0A0L0C3M6 K1Q3V6 A0A0B6ZQ45 A0A023F4V6 A0A0N8A8E5 F7DFI1 B1H2T4 G3TWX8 A0A0N8DV92 A0A0P6H6W2
EC Number
6.3.5.1
Pubmed
19121390
26354079
22118469
25315136
18362917
19820115
+ More
17994087 15632085 24845553 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 18327897 27448710 20075255 25830018 28648823 18057021 21347285 21719571 21282665 20798317 24508170 20566863 28004739 25244985 24438588 25576852 23537049 25765539 12364791 14747013 17210077 17510324 27762356 26108605 22992520 25474469 18464734 20431018
17994087 15632085 24845553 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 18327897 27448710 20075255 25830018 28648823 18057021 21347285 21719571 21282665 20798317 24508170 20566863 28004739 25244985 24438588 25576852 23537049 25765539 12364791 14747013 17210077 17510324 27762356 26108605 22992520 25474469 18464734 20431018
EMBL
BABH01000660
BABH01000661
BABH01000662
KQ460761
KPJ12571.1
KQ459299
+ More
KPJ01899.1 RSAL01000021 RVE52525.1 ODYU01007314 SOQ49988.1 ODYU01002110 SOQ39197.1 NWSH01000547 PCG75736.1 AGBW02011630 OWR46407.1 GFDL01001311 JAV33734.1 GFDL01001298 JAV33747.1 UFQS01000799 UFQT01000654 UFQT01000799 SSX06993.1 SSX26169.1 KA648500 AFP63129.1 CVRI01000019 CRK90414.1 KQ971338 EFA01802.1 NEVH01000597 PNF43572.1 CH916370 EDW00290.1 CH379064 EAL31647.1 CH479197 EDW27727.1 CP012528 ALC49803.1 KK852872 KDR14569.1 OUUW01000003 SPP78086.1 GAKP01019949 GAKP01019948 GAKP01019947 JAC39005.1 CH964239 EDW82448.1 CM000366 EDX17847.1 GDHF01005542 GDHF01003534 JAI46772.1 JAI48780.1 CM000162 EDX01609.1 CH480835 EDW48787.1 BT088846 AE014298 ACS68163.1 AHN59694.1 BT001474 GBXI01005411 JAD08881.1 NNAY01001171 OXU24892.1 CH902625 EDV35146.1 CH933813 EDW10989.1 CH940651 KRF82014.1 EDW65111.1 KRF82015.1 CH954180 EDV47169.1 KQ976396 KYM92874.1 ADTU01011806 KQ978448 KYM93946.1 KQ414614 KOC68889.1 KZ288222 PBC32056.1 GL888417 EGI61203.1 KQ979973 KYN18379.1 GL768788 EFZ11008.1 GL444981 EFN60322.1 KK107087 EZA59845.1 CCAG010007749 KQ981727 KYN37081.1 JXJN01028107 JXJN01028108 KQ434820 KZC06994.1 DS235363 EEB15245.1 GEZM01080274 JAV62234.1 KQ768821 OAD53069.1 GEBQ01025660 JAT14317.1 GECU01006253 JAT01454.1 KQ983211 KYQ46663.1 AXCN02002022 AXCN02002023 AXCM01008589 GL448193 EFN85146.1 ATLV01023368 KE525342 KFB48993.1 GDRN01048570 JAI66644.1 GACK01006857 JAA58177.1 KB632168 ERL89342.1 APCN01003987 APCN01003988 GANP01002931 JAB81537.1 KK115914 KFM66357.1 GGLE01001616 MBY05742.1 AAAB01008846 EAA06345.5 CH477497 EAT39937.1 KQ435720 KOX78680.1 CM004472 OCT83757.1 JRES01000952 KNC26852.1 JH815808 EKC23525.1 HACG01023066 CEK69931.1 GBBI01002659 JAC16053.1 GDIP01172377 JAJ51025.1 AAPN01149491 AAPN01149492 AAMC01029887 AAMC01029888 BC161121 AAI61121.1 GDIQ01088532 JAN06205.1 GDIQ01036305 JAN58432.1
KPJ01899.1 RSAL01000021 RVE52525.1 ODYU01007314 SOQ49988.1 ODYU01002110 SOQ39197.1 NWSH01000547 PCG75736.1 AGBW02011630 OWR46407.1 GFDL01001311 JAV33734.1 GFDL01001298 JAV33747.1 UFQS01000799 UFQT01000654 UFQT01000799 SSX06993.1 SSX26169.1 KA648500 AFP63129.1 CVRI01000019 CRK90414.1 KQ971338 EFA01802.1 NEVH01000597 PNF43572.1 CH916370 EDW00290.1 CH379064 EAL31647.1 CH479197 EDW27727.1 CP012528 ALC49803.1 KK852872 KDR14569.1 OUUW01000003 SPP78086.1 GAKP01019949 GAKP01019948 GAKP01019947 JAC39005.1 CH964239 EDW82448.1 CM000366 EDX17847.1 GDHF01005542 GDHF01003534 JAI46772.1 JAI48780.1 CM000162 EDX01609.1 CH480835 EDW48787.1 BT088846 AE014298 ACS68163.1 AHN59694.1 BT001474 GBXI01005411 JAD08881.1 NNAY01001171 OXU24892.1 CH902625 EDV35146.1 CH933813 EDW10989.1 CH940651 KRF82014.1 EDW65111.1 KRF82015.1 CH954180 EDV47169.1 KQ976396 KYM92874.1 ADTU01011806 KQ978448 KYM93946.1 KQ414614 KOC68889.1 KZ288222 PBC32056.1 GL888417 EGI61203.1 KQ979973 KYN18379.1 GL768788 EFZ11008.1 GL444981 EFN60322.1 KK107087 EZA59845.1 CCAG010007749 KQ981727 KYN37081.1 JXJN01028107 JXJN01028108 KQ434820 KZC06994.1 DS235363 EEB15245.1 GEZM01080274 JAV62234.1 KQ768821 OAD53069.1 GEBQ01025660 JAT14317.1 GECU01006253 JAT01454.1 KQ983211 KYQ46663.1 AXCN02002022 AXCN02002023 AXCM01008589 GL448193 EFN85146.1 ATLV01023368 KE525342 KFB48993.1 GDRN01048570 JAI66644.1 GACK01006857 JAA58177.1 KB632168 ERL89342.1 APCN01003987 APCN01003988 GANP01002931 JAB81537.1 KK115914 KFM66357.1 GGLE01001616 MBY05742.1 AAAB01008846 EAA06345.5 CH477497 EAT39937.1 KQ435720 KOX78680.1 CM004472 OCT83757.1 JRES01000952 KNC26852.1 JH815808 EKC23525.1 HACG01023066 CEK69931.1 GBBI01002659 JAC16053.1 GDIP01172377 JAJ51025.1 AAPN01149491 AAPN01149492 AAMC01029887 AAMC01029888 BC161121 AAI61121.1 GDIQ01088532 JAN06205.1 GDIQ01036305 JAN58432.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000218220
UP000007151
+ More
UP000095301 UP000183832 UP000007266 UP000095300 UP000235965 UP000001070 UP000192223 UP000001819 UP000008744 UP000092553 UP000027135 UP000268350 UP000007798 UP000000304 UP000002282 UP000001292 UP000000803 UP000192221 UP000002358 UP000215335 UP000007801 UP000009192 UP000008792 UP000008711 UP000078540 UP000005203 UP000005205 UP000078542 UP000053825 UP000242457 UP000007755 UP000078492 UP000000311 UP000053097 UP000092444 UP000078541 UP000092445 UP000091820 UP000078200 UP000092443 UP000092460 UP000076502 UP000009046 UP000075809 UP000075886 UP000075883 UP000076408 UP000008237 UP000075920 UP000075900 UP000030765 UP000075881 UP000075884 UP000075880 UP000030742 UP000075840 UP000075902 UP000076407 UP000054359 UP000075903 UP000085678 UP000007062 UP000008820 UP000053105 UP000186698 UP000037069 UP000005408 UP000002279 UP000008143 UP000007646
UP000095301 UP000183832 UP000007266 UP000095300 UP000235965 UP000001070 UP000192223 UP000001819 UP000008744 UP000092553 UP000027135 UP000268350 UP000007798 UP000000304 UP000002282 UP000001292 UP000000803 UP000192221 UP000002358 UP000215335 UP000007801 UP000009192 UP000008792 UP000008711 UP000078540 UP000005203 UP000005205 UP000078542 UP000053825 UP000242457 UP000007755 UP000078492 UP000000311 UP000053097 UP000092444 UP000078541 UP000092445 UP000091820 UP000078200 UP000092443 UP000092460 UP000076502 UP000009046 UP000075809 UP000075886 UP000075883 UP000076408 UP000008237 UP000075920 UP000075900 UP000030765 UP000075881 UP000075884 UP000075880 UP000030742 UP000075840 UP000075902 UP000076407 UP000054359 UP000075903 UP000085678 UP000007062 UP000008820 UP000053105 UP000186698 UP000037069 UP000005408 UP000002279 UP000008143 UP000007646
Interpro
SUPFAM
SSF56317
SSF56317
Gene 3D
ProteinModelPortal
H9IV80
A0A194R9P4
A0A194Q906
A0A3S2M6H4
A0A2H1WA98
A0A2H1VEG0
+ More
A0A2A4JVD5 A0A212EY75 A0A1Q3G1P3 A0A1Q3G1Q7 A0A336KV63 T1PKA9 A0A1J1HV96 D1ZZT1 A0A1I8NS65 A0A2J7RRY1 B4JL37 A0A1W4XJK8 Q29HW0 B4GY71 A0A0M3QZQ0 A0A067R837 A0A3B0JXK5 A0A034VB33 B4NC89 B4R4E5 A0A0K8W6T7 B4Q2E8 B4IG53 C5WLN1 Q9VYA0 A0A1W4VGV4 K7IZ08 A0A0A1XD61 A0A232F2S6 B3MW01 B4L7J5 A0A0Q9WMA9 B4M2T6 B3NWK7 A0A195BW65 A0A088AIB9 A0A158NCC5 A0A151I6Z7 A0A0L7RDJ9 A0A2A3ELI5 F4WX79 A0A151J5W2 E9J843 E2B1W6 A0A026WUW9 A0A1B0G4F1 A0A195F9M6 A0A1B0AHA3 A0A1A9WFA1 A0A1A9VWF6 A0A1A9YF37 A0A1B0C7C8 A0A154P6M5 E0VPD9 A0A1Y1KLN7 A0A310SFQ0 A0A1B6KSA4 A0A1B6JRC4 A0A151WFS0 A0A182QC98 A0A182MPM7 A0A182YQW1 E2BGL8 A0A182W416 A0A182RVT2 A0A084WFJ9 A0A182KGG7 A0A0P4WDD5 A0A182N052 A0A182JFP2 L7M2U8 U4U8I8 A0A182HIJ6 A0A182U6G3 A0A182WY73 V5HYE3 A0A087TML8 A0A182VMG5 A0A1S3JZK0 A0A2R5L8C7 Q7PS02 Q16Z66 A0A0M9A797 A0A1L8GIT4 A0A0L0C3M6 K1Q3V6 A0A0B6ZQ45 A0A023F4V6 A0A0N8A8E5 F7DFI1 B1H2T4 G3TWX8 A0A0N8DV92 A0A0P6H6W2
A0A2A4JVD5 A0A212EY75 A0A1Q3G1P3 A0A1Q3G1Q7 A0A336KV63 T1PKA9 A0A1J1HV96 D1ZZT1 A0A1I8NS65 A0A2J7RRY1 B4JL37 A0A1W4XJK8 Q29HW0 B4GY71 A0A0M3QZQ0 A0A067R837 A0A3B0JXK5 A0A034VB33 B4NC89 B4R4E5 A0A0K8W6T7 B4Q2E8 B4IG53 C5WLN1 Q9VYA0 A0A1W4VGV4 K7IZ08 A0A0A1XD61 A0A232F2S6 B3MW01 B4L7J5 A0A0Q9WMA9 B4M2T6 B3NWK7 A0A195BW65 A0A088AIB9 A0A158NCC5 A0A151I6Z7 A0A0L7RDJ9 A0A2A3ELI5 F4WX79 A0A151J5W2 E9J843 E2B1W6 A0A026WUW9 A0A1B0G4F1 A0A195F9M6 A0A1B0AHA3 A0A1A9WFA1 A0A1A9VWF6 A0A1A9YF37 A0A1B0C7C8 A0A154P6M5 E0VPD9 A0A1Y1KLN7 A0A310SFQ0 A0A1B6KSA4 A0A1B6JRC4 A0A151WFS0 A0A182QC98 A0A182MPM7 A0A182YQW1 E2BGL8 A0A182W416 A0A182RVT2 A0A084WFJ9 A0A182KGG7 A0A0P4WDD5 A0A182N052 A0A182JFP2 L7M2U8 U4U8I8 A0A182HIJ6 A0A182U6G3 A0A182WY73 V5HYE3 A0A087TML8 A0A182VMG5 A0A1S3JZK0 A0A2R5L8C7 Q7PS02 Q16Z66 A0A0M9A797 A0A1L8GIT4 A0A0L0C3M6 K1Q3V6 A0A0B6ZQ45 A0A023F4V6 A0A0N8A8E5 F7DFI1 B1H2T4 G3TWX8 A0A0N8DV92 A0A0P6H6W2
PDB
3ILV
E-value=5.20154e-39,
Score=405
Ontologies
PATHWAY
GO
PANTHER
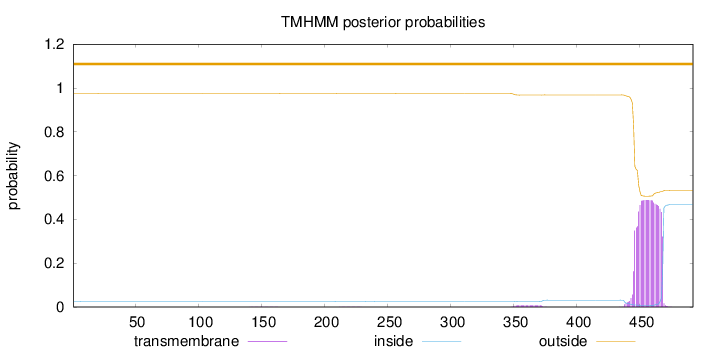
Topology
Length:
492
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.97679
Exp number, first 60 AAs:
0.00125
Total prob of N-in:
0.02575
outside
1 - 492
Population Genetic Test Statistics
Pi
191.531244
Theta
161.505795
Tajima's D
0.010191
CLR
1.036543
CSRT
0.373831308434578
Interpretation
Uncertain
