Gene
KWMTBOMO07947
Pre Gene Modal
BGIBMGA001160
Annotation
PREDICTED:_probable_glutamine-dependent_NAD(+)_synthetase_[Papilio_xuthus]
Full name
Glutamine-dependent NAD(+) synthetase
Alternative Name
NAD(+) synthase [glutamine-hydrolyzing]
NAD(+) synthetase
NAD(+) synthetase
Location in the cell
Extracellular Reliability : 1.757
Sequence
CDS
ATGGTTCTGTCGTATTTGTTCGCGCAGCTAATGCTCTGGGTCCGCGGCAGACCTGGAGGTCTTTTAGTACTGGGATCCTCAAACGTCGACGAAGCCCTGCGAGGCTACATGACCAAGTACGATTGTTCCAGTGCAGATATTAATCCCATCGGTGGAATATCTAAGACTGACCTCAAATCTTTCTTACATTACGCTAAGAATAGATTTTTCTTGCCGTCTCTATCGGAAATTCTAGAAGCACCTCCGACTGCAGAACTCGAGCCATTGGCCGACGGTCAGATAACGCAAACCGACGAACAAGACATGGGTATGACATACGCTGAACTGTCCGAATTCGGAACTTTAAGGAAGACCTATAAATGCGGCCCTTACAGCATGTTCGAGAAACTGGTTCATAAGTGGAGTGATAAATGTACGCCTAAGGAGGTGGCTGAAAAGGTGAAACATTTTTTCCGGTGTTACGCGATCAACAGGCACAAGATGACGGTGCTGACGCCTTCCTACCACGCCGAGACATACAGCCCCGACGACAACCGCTTCGATCTCAGACCTTTCCTATATCGAGTACACTGGAATTGGCAGTTTAAGACCATAGATGACGCTGTCATACAAATGAGCAAGGAAAAGCGTACATCAGTTAGCCGGGTTGGATATGAACAGAGCAAAACAAATCCGAATTCTCCGACTACTACATTGAGTTCGTCGTTCACAAAGAACGACAGACAGGGTGTACTGATATGA
Protein
MVLSYLFAQLMLWVRGRPGGLLVLGSSNVDEALRGYMTKYDCSSADINPIGGISKTDLKSFLHYAKNRFFLPSLSEILEAPPTAELEPLADGQITQTDEQDMGMTYAELSEFGTLRKTYKCGPYSMFEKLVHKWSDKCTPKEVAEKVKHFFRCYAINRHKMTVLTPSYHAETYSPDDNRFDLRPFLYRVHWNWQFKTIDDAVIQMSKEKRTSVSRVGYEQSKTNPNSPTTTLSSSFTKNDRQGVLI
Summary
Description
Catalyzes the ATP-dependent amidation of deamido-NAD to form NAD. Uses L-glutamine as a nitrogen source.
Catalytic Activity
ATP + deamido-NAD(+) + H2O + L-glutamine = AMP + diphosphate + H(+) + L-glutamate + NAD(+)
Similarity
In the C-terminal section; belongs to the NAD synthetase family.
Keywords
ATP-binding
Complete proteome
Ligase
NAD
Nucleotide-binding
Reference proteome
Feature
chain Glutamine-dependent NAD(+) synthetase
Uniprot
H9IV80
A0A2H1WA98
A0A2H1VEG0
A0A2A4JVD5
A0A194Q906
A0A3S2M6H4
+ More
A0A212EY75 A0A194R9P4 A0A1B6GM02 A0A1B6JE34 D1ZZT1 A0A1B6KSA4 A0A067R837 A0A2J7RRV5 A0A182GI80 A0A034VB33 A0A2J7RRY1 A0A336KV63 A0A0K8W6T7 A0A1J1HV96 U4U8I8 A0A0A1XD61 A0A1W4XJK8 A0A1B6D2S2 A0A1B6D5S3 A0A1Y1KLN7 A0A1Q3G1Q7 A0A2P8ZCY9 T1PKA9 A0A1I8NS65 A0A1Q3G1P3 V9KIP6 Q16E59 Q16Z66 A0A0Q9WMA9 B4M2T6 A0A1S3CXU3 A0A1A8Q9N7 A0A182TC16 B4JL37 A0A0M3QZQ0 A0A1A8A7H0 A0A3B3XLH7 U3I5Q1 A0A1A8L7W3 E0VPD9 U3K2E3 B3MW01 G1MU36 B4L7J5 A0A218URA9 A0A3B0JXK5 F1P4D3 A0A2M4AMM0 A0A091JTW6 A0A1L8GIT4 W4XND2 A0A1A9WFA1 B4Q2E8 A0A2J8S6C0 A0A1W4VGV4 A0A0J7KXT5 B3NWK7 A0A2J8S689 Q29HW0 B4GY71 A0A1A9YF37 A0A1B0C7C8 B4NC89 B1H2T4 A0A099ZVA5 A0A2K6SEN9 A0A182N052 A0A0B4J216 A0A2P4T031 R0L011 A0A2I4AND5 A0A3P4LZZ1 A0A091QST8 A0A087VMG6 G3VZL4 Q5ZMA6 A0A3Q3FHF9 A0A2J8IQ76 C3Y7L9 A0A3Q3JFD8 A0A093NCQ3 A0A226P2X0 G1PXJ9 H0Z4H8 A0A0A0AK77 A0A1B0AHA3 A0A2J8S6A6 A0A1A9VWF6 A0A091VB37 M7B7J6 A0A212DGM2 A0A093QM28 A0A182JFP2 A0A087XA41
A0A212EY75 A0A194R9P4 A0A1B6GM02 A0A1B6JE34 D1ZZT1 A0A1B6KSA4 A0A067R837 A0A2J7RRV5 A0A182GI80 A0A034VB33 A0A2J7RRY1 A0A336KV63 A0A0K8W6T7 A0A1J1HV96 U4U8I8 A0A0A1XD61 A0A1W4XJK8 A0A1B6D2S2 A0A1B6D5S3 A0A1Y1KLN7 A0A1Q3G1Q7 A0A2P8ZCY9 T1PKA9 A0A1I8NS65 A0A1Q3G1P3 V9KIP6 Q16E59 Q16Z66 A0A0Q9WMA9 B4M2T6 A0A1S3CXU3 A0A1A8Q9N7 A0A182TC16 B4JL37 A0A0M3QZQ0 A0A1A8A7H0 A0A3B3XLH7 U3I5Q1 A0A1A8L7W3 E0VPD9 U3K2E3 B3MW01 G1MU36 B4L7J5 A0A218URA9 A0A3B0JXK5 F1P4D3 A0A2M4AMM0 A0A091JTW6 A0A1L8GIT4 W4XND2 A0A1A9WFA1 B4Q2E8 A0A2J8S6C0 A0A1W4VGV4 A0A0J7KXT5 B3NWK7 A0A2J8S689 Q29HW0 B4GY71 A0A1A9YF37 A0A1B0C7C8 B4NC89 B1H2T4 A0A099ZVA5 A0A2K6SEN9 A0A182N052 A0A0B4J216 A0A2P4T031 R0L011 A0A2I4AND5 A0A3P4LZZ1 A0A091QST8 A0A087VMG6 G3VZL4 Q5ZMA6 A0A3Q3FHF9 A0A2J8IQ76 C3Y7L9 A0A3Q3JFD8 A0A093NCQ3 A0A226P2X0 G1PXJ9 H0Z4H8 A0A0A0AK77 A0A1B0AHA3 A0A2J8S6A6 A0A1A9VWF6 A0A091VB37 M7B7J6 A0A212DGM2 A0A093QM28 A0A182JFP2 A0A087XA41
EC Number
6.3.5.1
Pubmed
19121390
26354079
22118469
18362917
19820115
24845553
+ More
26483478 25348373 23537049 25830018 28004739 29403074 25315136 24402279 17510324 17994087 18057021 23749191 20566863 20838655 15592404 27762356 17550304 15632085 20431018 16554811 21269460 21709235 15642098 18563158 24621616 21993624 20360741 23624526 29294181
26483478 25348373 23537049 25830018 28004739 29403074 25315136 24402279 17510324 17994087 18057021 23749191 20566863 20838655 15592404 27762356 17550304 15632085 20431018 16554811 21269460 21709235 15642098 18563158 24621616 21993624 20360741 23624526 29294181
EMBL
BABH01000660
BABH01000661
BABH01000662
ODYU01007314
SOQ49988.1
ODYU01002110
+ More
SOQ39197.1 NWSH01000547 PCG75736.1 KQ459299 KPJ01899.1 RSAL01000021 RVE52525.1 AGBW02011630 OWR46407.1 KQ460761 KPJ12571.1 GECZ01006416 JAS63353.1 GECU01010483 JAS97223.1 KQ971338 EFA01802.1 GEBQ01025660 JAT14317.1 KK852872 KDR14569.1 NEVH01000597 PNF43570.1 JXUM01012269 KQ560350 KXJ82898.1 GAKP01019949 GAKP01019948 GAKP01019947 JAC39005.1 PNF43572.1 UFQS01000799 UFQT01000654 UFQT01000799 SSX06993.1 SSX26169.1 GDHF01005542 GDHF01003534 JAI46772.1 JAI48780.1 CVRI01000019 CRK90414.1 KB632168 ERL89342.1 GBXI01005411 JAD08881.1 GEDC01017318 JAS19980.1 GEDC01016266 JAS21032.1 GEZM01080274 JAV62234.1 GFDL01001298 JAV33747.1 PYGN01000098 PSN54369.1 KA648500 AFP63129.1 GFDL01001311 JAV33734.1 JW865750 AFO98267.1 CH479099 EAT32516.1 CH477497 EAT39937.1 CH940651 KRF82014.1 EDW65111.1 KRF82015.1 HAEG01011498 SBR89914.1 CH916370 EDW00290.1 CP012528 ALC49803.1 HADY01011655 SBP50140.1 ADON01092589 HAEF01003340 SBR40722.1 DS235363 EEB15245.1 AGTO01002176 CH902625 EDV35146.1 CH933813 EDW10989.1 MUZQ01000167 OWK56176.1 OUUW01000003 SPP78086.1 AADN05000685 GGFK01008651 MBW41972.1 KK531466 KFP28160.1 CM004472 OCT83757.1 AAGJ04123609 CM000162 EDX01609.1 NDHI03003604 PNJ16294.1 LBMM01002037 KMQ95367.1 CH954180 EDV47169.1 PNJ16295.1 CH379064 EAL31647.1 CH479197 EDW27727.1 JXJN01028107 JXJN01028108 CH964239 EDW82448.1 AAMC01029887 AAMC01029888 BC161121 AAI61121.1 KL897677 KGL84745.1 AP000867 AP002387 PPHD01014277 POI29711.1 KB743402 EOA98928.1 CYRY02003651 VCW68246.1 KK680580 KFQ12454.1 KL498683 KFO13808.1 AEFK01224255 AJ719478 NBAG03000638 PNI12665.1 GG666489 EEN63847.1 KL224691 KFW62678.1 AWGT02000180 OXB74375.1 AAPE02059498 AAPE02059499 ABQF01001419 KL872041 KGL94317.1 PNJ16296.1 KK422457 KFQ86903.1 KB565340 EMP28143.1 MKHE01000002 OWK17331.1 KL672977 KFW87460.1 AYCK01026215 AYCK01026216 AYCK01026217
SOQ39197.1 NWSH01000547 PCG75736.1 KQ459299 KPJ01899.1 RSAL01000021 RVE52525.1 AGBW02011630 OWR46407.1 KQ460761 KPJ12571.1 GECZ01006416 JAS63353.1 GECU01010483 JAS97223.1 KQ971338 EFA01802.1 GEBQ01025660 JAT14317.1 KK852872 KDR14569.1 NEVH01000597 PNF43570.1 JXUM01012269 KQ560350 KXJ82898.1 GAKP01019949 GAKP01019948 GAKP01019947 JAC39005.1 PNF43572.1 UFQS01000799 UFQT01000654 UFQT01000799 SSX06993.1 SSX26169.1 GDHF01005542 GDHF01003534 JAI46772.1 JAI48780.1 CVRI01000019 CRK90414.1 KB632168 ERL89342.1 GBXI01005411 JAD08881.1 GEDC01017318 JAS19980.1 GEDC01016266 JAS21032.1 GEZM01080274 JAV62234.1 GFDL01001298 JAV33747.1 PYGN01000098 PSN54369.1 KA648500 AFP63129.1 GFDL01001311 JAV33734.1 JW865750 AFO98267.1 CH479099 EAT32516.1 CH477497 EAT39937.1 CH940651 KRF82014.1 EDW65111.1 KRF82015.1 HAEG01011498 SBR89914.1 CH916370 EDW00290.1 CP012528 ALC49803.1 HADY01011655 SBP50140.1 ADON01092589 HAEF01003340 SBR40722.1 DS235363 EEB15245.1 AGTO01002176 CH902625 EDV35146.1 CH933813 EDW10989.1 MUZQ01000167 OWK56176.1 OUUW01000003 SPP78086.1 AADN05000685 GGFK01008651 MBW41972.1 KK531466 KFP28160.1 CM004472 OCT83757.1 AAGJ04123609 CM000162 EDX01609.1 NDHI03003604 PNJ16294.1 LBMM01002037 KMQ95367.1 CH954180 EDV47169.1 PNJ16295.1 CH379064 EAL31647.1 CH479197 EDW27727.1 JXJN01028107 JXJN01028108 CH964239 EDW82448.1 AAMC01029887 AAMC01029888 BC161121 AAI61121.1 KL897677 KGL84745.1 AP000867 AP002387 PPHD01014277 POI29711.1 KB743402 EOA98928.1 CYRY02003651 VCW68246.1 KK680580 KFQ12454.1 KL498683 KFO13808.1 AEFK01224255 AJ719478 NBAG03000638 PNI12665.1 GG666489 EEN63847.1 KL224691 KFW62678.1 AWGT02000180 OXB74375.1 AAPE02059498 AAPE02059499 ABQF01001419 KL872041 KGL94317.1 PNJ16296.1 KK422457 KFQ86903.1 KB565340 EMP28143.1 MKHE01000002 OWK17331.1 KL672977 KFW87460.1 AYCK01026215 AYCK01026216 AYCK01026217
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
+ More
UP000007266 UP000027135 UP000235965 UP000069940 UP000249989 UP000183832 UP000030742 UP000192223 UP000245037 UP000095301 UP000095300 UP000008820 UP000008792 UP000079169 UP000075901 UP000001070 UP000092553 UP000261480 UP000016666 UP000009046 UP000016665 UP000007801 UP000001645 UP000009192 UP000197619 UP000268350 UP000000539 UP000186698 UP000007110 UP000091820 UP000002282 UP000192221 UP000036403 UP000008711 UP000001819 UP000008744 UP000092443 UP000092460 UP000007798 UP000008143 UP000053641 UP000233220 UP000075884 UP000005640 UP000192220 UP000007648 UP000261660 UP000001554 UP000261600 UP000054081 UP000198419 UP000001074 UP000007754 UP000053858 UP000092445 UP000078200 UP000031443 UP000053258 UP000075880 UP000028760
UP000007266 UP000027135 UP000235965 UP000069940 UP000249989 UP000183832 UP000030742 UP000192223 UP000245037 UP000095301 UP000095300 UP000008820 UP000008792 UP000079169 UP000075901 UP000001070 UP000092553 UP000261480 UP000016666 UP000009046 UP000016665 UP000007801 UP000001645 UP000009192 UP000197619 UP000268350 UP000000539 UP000186698 UP000007110 UP000091820 UP000002282 UP000192221 UP000036403 UP000008711 UP000001819 UP000008744 UP000092443 UP000092460 UP000007798 UP000008143 UP000053641 UP000233220 UP000075884 UP000005640 UP000192220 UP000007648 UP000261660 UP000001554 UP000261600 UP000054081 UP000198419 UP000001074 UP000007754 UP000053858 UP000092445 UP000078200 UP000031443 UP000053258 UP000075880 UP000028760
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IV80
A0A2H1WA98
A0A2H1VEG0
A0A2A4JVD5
A0A194Q906
A0A3S2M6H4
+ More
A0A212EY75 A0A194R9P4 A0A1B6GM02 A0A1B6JE34 D1ZZT1 A0A1B6KSA4 A0A067R837 A0A2J7RRV5 A0A182GI80 A0A034VB33 A0A2J7RRY1 A0A336KV63 A0A0K8W6T7 A0A1J1HV96 U4U8I8 A0A0A1XD61 A0A1W4XJK8 A0A1B6D2S2 A0A1B6D5S3 A0A1Y1KLN7 A0A1Q3G1Q7 A0A2P8ZCY9 T1PKA9 A0A1I8NS65 A0A1Q3G1P3 V9KIP6 Q16E59 Q16Z66 A0A0Q9WMA9 B4M2T6 A0A1S3CXU3 A0A1A8Q9N7 A0A182TC16 B4JL37 A0A0M3QZQ0 A0A1A8A7H0 A0A3B3XLH7 U3I5Q1 A0A1A8L7W3 E0VPD9 U3K2E3 B3MW01 G1MU36 B4L7J5 A0A218URA9 A0A3B0JXK5 F1P4D3 A0A2M4AMM0 A0A091JTW6 A0A1L8GIT4 W4XND2 A0A1A9WFA1 B4Q2E8 A0A2J8S6C0 A0A1W4VGV4 A0A0J7KXT5 B3NWK7 A0A2J8S689 Q29HW0 B4GY71 A0A1A9YF37 A0A1B0C7C8 B4NC89 B1H2T4 A0A099ZVA5 A0A2K6SEN9 A0A182N052 A0A0B4J216 A0A2P4T031 R0L011 A0A2I4AND5 A0A3P4LZZ1 A0A091QST8 A0A087VMG6 G3VZL4 Q5ZMA6 A0A3Q3FHF9 A0A2J8IQ76 C3Y7L9 A0A3Q3JFD8 A0A093NCQ3 A0A226P2X0 G1PXJ9 H0Z4H8 A0A0A0AK77 A0A1B0AHA3 A0A2J8S6A6 A0A1A9VWF6 A0A091VB37 M7B7J6 A0A212DGM2 A0A093QM28 A0A182JFP2 A0A087XA41
A0A212EY75 A0A194R9P4 A0A1B6GM02 A0A1B6JE34 D1ZZT1 A0A1B6KSA4 A0A067R837 A0A2J7RRV5 A0A182GI80 A0A034VB33 A0A2J7RRY1 A0A336KV63 A0A0K8W6T7 A0A1J1HV96 U4U8I8 A0A0A1XD61 A0A1W4XJK8 A0A1B6D2S2 A0A1B6D5S3 A0A1Y1KLN7 A0A1Q3G1Q7 A0A2P8ZCY9 T1PKA9 A0A1I8NS65 A0A1Q3G1P3 V9KIP6 Q16E59 Q16Z66 A0A0Q9WMA9 B4M2T6 A0A1S3CXU3 A0A1A8Q9N7 A0A182TC16 B4JL37 A0A0M3QZQ0 A0A1A8A7H0 A0A3B3XLH7 U3I5Q1 A0A1A8L7W3 E0VPD9 U3K2E3 B3MW01 G1MU36 B4L7J5 A0A218URA9 A0A3B0JXK5 F1P4D3 A0A2M4AMM0 A0A091JTW6 A0A1L8GIT4 W4XND2 A0A1A9WFA1 B4Q2E8 A0A2J8S6C0 A0A1W4VGV4 A0A0J7KXT5 B3NWK7 A0A2J8S689 Q29HW0 B4GY71 A0A1A9YF37 A0A1B0C7C8 B4NC89 B1H2T4 A0A099ZVA5 A0A2K6SEN9 A0A182N052 A0A0B4J216 A0A2P4T031 R0L011 A0A2I4AND5 A0A3P4LZZ1 A0A091QST8 A0A087VMG6 G3VZL4 Q5ZMA6 A0A3Q3FHF9 A0A2J8IQ76 C3Y7L9 A0A3Q3JFD8 A0A093NCQ3 A0A226P2X0 G1PXJ9 H0Z4H8 A0A0A0AK77 A0A1B0AHA3 A0A2J8S6A6 A0A1A9VWF6 A0A091VB37 M7B7J6 A0A212DGM2 A0A093QM28 A0A182JFP2 A0A087XA41
PDB
3ILV
E-value=8.89892e-11,
Score=158
Ontologies
PANTHER
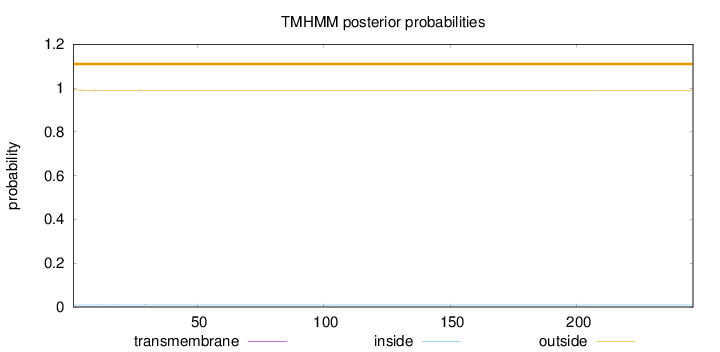
Topology
Length:
246
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02072
Exp number, first 60 AAs:
0.02072
Total prob of N-in:
0.00942
outside
1 - 246
Population Genetic Test Statistics
Pi
228.827833
Theta
177.704315
Tajima's D
1.168503
CLR
0
CSRT
0.706564671766412
Interpretation
Uncertain
