Gene
KWMTBOMO07939
Pre Gene Modal
BGIBMGA000983
Annotation
PREDICTED:_TWiK_family_of_potassium_channels_protein_7_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.616
Sequence
CDS
ATGTGTAAAAATGAGAAGAACGTGTCATGGGAGACGGAGGCACTTATGGGTGCAAAGCCTGAATCGAGATGGAGCAATTTCTGTCCAAGAGAAGCCTTGTCCCATGTTGGGTTATTCGTTGCGTTAATGCTGTACACAGCTGGCGGCGGATTGGTATTCCGTGCGTTAGAGTATCCAGCTGAAATGGCTAAACAGGATTTTCATAGGGAACGTGTTCTAAGTGAAAGATGGAATTTAATACGTTTTGTAGCTCAATGGACAAACGGGACTGAGTATTTTAATGGCGTTGAAGAGCTGCTAAGCCATCATTTAGCTATTTACGAAAAGGTGTTAGAAGAAGCGTCATCAGATGGCATATCGTTGGAAGTAGAAAAGAATTTTCCACCAACCGAAGAAAGATGGAGCATTTTGCAAGCTGTATTTTTTTCTTCAACAGTGCTCACTACTATAGGTTACGGTAACATAGTGCCGGTAACATTTTGGGGAAGGTTGTTTTGTATCGCTTACGCCTTGATAGGAATACCATTCACATTAACGGTCATCGCTGATCTAGGCAGAGTTTTCGCCACATGTGTTTCCTTTATCGCTAAACATCTACCGTCACCACCTAGTAAGTTAACATGA
Protein
MCKNEKNVSWETEALMGAKPESRWSNFCPREALSHVGLFVALMLYTAGGGLVFRALEYPAEMAKQDFHRERVLSERWNLIRFVAQWTNGTEYFNGVEELLSHHLAIYEKVLEEASSDGISLEVEKNFPPTEERWSILQAVFFSSTVLTTIGYGNIVPVTFWGRLFCIAYALIGIPFTLTVIADLGRVFATCVSFIAKHLPSPPSKLT
Summary
Similarity
Belongs to the two pore domain potassium channel (TC 1.A.1.8) family.
Uniprot
A0A2H1VT12
A0A3S2LVT4
H9IUQ3
A0A2W1BYK4
A0A2A4J5D2
A0A0N0PDK3
+ More
A0A194Q8P4 A0A1I8N2A9 A0A0Q9WN14 B4L1K4 Q29HS6 B4GYD9 Q9VYY5 B4IK18 B4R343 B4PYA6 W8AP75 A0A0L0CKA9 B4MSC4 B3NUD1 A0A3B0KEY0 A0A1W4V1W9 B4M355 A0A0K8W0C5 B3MWJ0 A0A0A1WJD1 A0A084VQ45 U5EEQ4 A0A034WRL0 A0A182FMB5 A0A182YH38 W5JI18 Q17DS8 B0XGP9 A0A0M3QZ46 A0A139WIX4 E2B1X3 A0A0K8U275 A0A1Y1KR28 A0A1A9VBQ4 A0A1B0G8B4 A0A1A9Z634 A0A1B0EXA1 A0A182PUM2 U4UHK3 A0A182JKC3 N6UUV7 A0A1A9XXQ9 A0A1B0APF7 A0A182MF40 A0A0L7RDT6 Q7Q822 A7UTI7 A0A182I658 A0A154P4Y7 A0A182W5Z7 A0A088AIB6 A0A2A3ELB4 A0A232F9I3 A0A310S628 A0A0M9A8K7 A0A067QW29 A0A182RVW7 A0A1J1HR05 A0A088ATV4 A0A195CEI0 A0A336KAZ3 A0A0P5I3V3 A0A0P4ZF17 A0A0P5NGR5 A0A0P5APG8 A0A0P5KRQ2 A0A0A9YQN9 A0A0A9XKG5 A0A146M548 A0A0P5SIA8 A0A0A9XF06 A0A0P5KW34 A0A0A9XQD1 A0A0P4YBJ1 A0A0N8B2D2 A0A0P5LTL6 A0A0P5JVA9 A0A0P5JI11 A0A0P6EVF7 A0A0P5T429 A0A0P5VSX9 A0A0P5RLL0 A0A0P5QMF1 A0A0P4XT46 A0A0P5GA70 A0A0P6F080 A0A0P5G0I6 A0A0P5DU13 A0A0P6I569 A0A0P6BV24 A0A0P5WD21 A0A0L7KQ02
A0A194Q8P4 A0A1I8N2A9 A0A0Q9WN14 B4L1K4 Q29HS6 B4GYD9 Q9VYY5 B4IK18 B4R343 B4PYA6 W8AP75 A0A0L0CKA9 B4MSC4 B3NUD1 A0A3B0KEY0 A0A1W4V1W9 B4M355 A0A0K8W0C5 B3MWJ0 A0A0A1WJD1 A0A084VQ45 U5EEQ4 A0A034WRL0 A0A182FMB5 A0A182YH38 W5JI18 Q17DS8 B0XGP9 A0A0M3QZ46 A0A139WIX4 E2B1X3 A0A0K8U275 A0A1Y1KR28 A0A1A9VBQ4 A0A1B0G8B4 A0A1A9Z634 A0A1B0EXA1 A0A182PUM2 U4UHK3 A0A182JKC3 N6UUV7 A0A1A9XXQ9 A0A1B0APF7 A0A182MF40 A0A0L7RDT6 Q7Q822 A7UTI7 A0A182I658 A0A154P4Y7 A0A182W5Z7 A0A088AIB6 A0A2A3ELB4 A0A232F9I3 A0A310S628 A0A0M9A8K7 A0A067QW29 A0A182RVW7 A0A1J1HR05 A0A088ATV4 A0A195CEI0 A0A336KAZ3 A0A0P5I3V3 A0A0P4ZF17 A0A0P5NGR5 A0A0P5APG8 A0A0P5KRQ2 A0A0A9YQN9 A0A0A9XKG5 A0A146M548 A0A0P5SIA8 A0A0A9XF06 A0A0P5KW34 A0A0A9XQD1 A0A0P4YBJ1 A0A0N8B2D2 A0A0P5LTL6 A0A0P5JVA9 A0A0P5JI11 A0A0P6EVF7 A0A0P5T429 A0A0P5VSX9 A0A0P5RLL0 A0A0P5QMF1 A0A0P4XT46 A0A0P5GA70 A0A0P6F080 A0A0P5G0I6 A0A0P5DU13 A0A0P6I569 A0A0P6BV24 A0A0P5WD21 A0A0L7KQ02
Pubmed
19121390
28756777
26354079
25315136
17994087
15632085
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 24495485 26108605 25830018 24438588 25348373 25244985 20920257 23761445 17510324 18362917 19820115 20798317 28004739 23537049 12364791 28648823 24845553 25401762 26823975 26227816
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 24495485 26108605 25830018 24438588 25348373 25244985 20920257 23761445 17510324 18362917 19820115 20798317 28004739 23537049 12364791 28648823 24845553 25401762 26823975 26227816
EMBL
ODYU01004270
SOQ43963.1
RSAL01000173
RVE45076.1
BABH01000681
KZ149944
+ More
PZC76853.1 NWSH01002968 PCG67181.1 KQ460202 KPJ17174.1 KQ459299 KPJ01907.1 CH933810 KRF94126.1 EDW07640.2 CH379064 EAL31681.3 CH479197 EDW27795.1 AE014298 AAF48048.2 CH480851 EDW51390.1 CM000366 EDX17645.1 CM000162 EDX01968.1 GAMC01018733 JAB87822.1 JRES01000290 KNC32695.1 CH963851 EDW75013.2 CH954180 EDV46046.1 OUUW01000011 SPP86880.1 CH940651 EDW65230.2 GDHF01007701 JAI44613.1 CH902625 EDV34975.2 GBXI01015531 GBXI01001155 JAC98760.1 JAD13137.1 ATLV01015123 KE525003 KFB40089.1 GANO01004214 JAB55657.1 GAKP01001965 JAC56987.1 ADMH02001125 ETN64032.1 CH477291 EAT44586.1 DS233053 EDS27772.1 CP012528 ALC48793.1 KQ971338 KYB27940.1 GL444981 EFN60329.1 GDHF01031698 JAI20616.1 GEZM01078763 JAV62630.1 CCAG010000229 AJVK01001016 AJVK01001017 KB632168 ERL89345.1 APGK01011346 APGK01011347 APGK01011348 APGK01011349 APGK01011350 APGK01011351 KB738451 ENN82602.1 JXJN01001399 AXCM01010835 KQ414614 KOC68896.1 AAAB01008948 EAA10378.2 EDO63978.1 APCN01007519 KQ434820 KZC06989.1 KZ288222 PBC32062.1 NNAY01000589 OXU27524.1 KQ768821 OAD53064.1 KQ435720 KOX78676.1 KK852878 KDR14501.1 CVRI01000019 CRK90411.1 KQ977873 KYM99115.1 UFQS01000116 UFQT01000116 SSW99953.1 SSX20333.1 GDIQ01218405 JAK33320.1 GDIP01213575 JAJ09827.1 GDIQ01145533 JAL06193.1 GDIP01196340 JAJ27062.1 GDIQ01180663 JAK71062.1 GBHO01009663 JAG33941.1 GBHO01024296 JAG19308.1 GDHC01003755 JAQ14874.1 GDIP01139549 JAL64165.1 GBHO01024292 JAG19312.1 GDIQ01179258 JAK72467.1 GBHO01024294 JAG19310.1 GDIP01229887 JAI93514.1 GDIQ01219603 JAK32122.1 GDIQ01165434 JAK86291.1 GDIQ01198408 JAK53317.1 GDIQ01199709 JAK52016.1 GDIQ01064758 JAN29979.1 GDIP01133636 JAL70078.1 GDIP01095804 JAM07911.1 GDIQ01099996 JAL51730.1 GDIQ01115197 JAL36529.1 GDIP01236936 JAI86465.1 GDIQ01251349 JAK00376.1 GDIQ01067606 JAN27131.1 GDIQ01248431 JAK03294.1 GDIP01151597 JAJ71805.1 GDIQ01011177 JAN83560.1 GDIP01010429 JAM93286.1 GDIP01087804 JAM15911.1 JTDY01007475 KOB65170.1
PZC76853.1 NWSH01002968 PCG67181.1 KQ460202 KPJ17174.1 KQ459299 KPJ01907.1 CH933810 KRF94126.1 EDW07640.2 CH379064 EAL31681.3 CH479197 EDW27795.1 AE014298 AAF48048.2 CH480851 EDW51390.1 CM000366 EDX17645.1 CM000162 EDX01968.1 GAMC01018733 JAB87822.1 JRES01000290 KNC32695.1 CH963851 EDW75013.2 CH954180 EDV46046.1 OUUW01000011 SPP86880.1 CH940651 EDW65230.2 GDHF01007701 JAI44613.1 CH902625 EDV34975.2 GBXI01015531 GBXI01001155 JAC98760.1 JAD13137.1 ATLV01015123 KE525003 KFB40089.1 GANO01004214 JAB55657.1 GAKP01001965 JAC56987.1 ADMH02001125 ETN64032.1 CH477291 EAT44586.1 DS233053 EDS27772.1 CP012528 ALC48793.1 KQ971338 KYB27940.1 GL444981 EFN60329.1 GDHF01031698 JAI20616.1 GEZM01078763 JAV62630.1 CCAG010000229 AJVK01001016 AJVK01001017 KB632168 ERL89345.1 APGK01011346 APGK01011347 APGK01011348 APGK01011349 APGK01011350 APGK01011351 KB738451 ENN82602.1 JXJN01001399 AXCM01010835 KQ414614 KOC68896.1 AAAB01008948 EAA10378.2 EDO63978.1 APCN01007519 KQ434820 KZC06989.1 KZ288222 PBC32062.1 NNAY01000589 OXU27524.1 KQ768821 OAD53064.1 KQ435720 KOX78676.1 KK852878 KDR14501.1 CVRI01000019 CRK90411.1 KQ977873 KYM99115.1 UFQS01000116 UFQT01000116 SSW99953.1 SSX20333.1 GDIQ01218405 JAK33320.1 GDIP01213575 JAJ09827.1 GDIQ01145533 JAL06193.1 GDIP01196340 JAJ27062.1 GDIQ01180663 JAK71062.1 GBHO01009663 JAG33941.1 GBHO01024296 JAG19308.1 GDHC01003755 JAQ14874.1 GDIP01139549 JAL64165.1 GBHO01024292 JAG19312.1 GDIQ01179258 JAK72467.1 GBHO01024294 JAG19310.1 GDIP01229887 JAI93514.1 GDIQ01219603 JAK32122.1 GDIQ01165434 JAK86291.1 GDIQ01198408 JAK53317.1 GDIQ01199709 JAK52016.1 GDIQ01064758 JAN29979.1 GDIP01133636 JAL70078.1 GDIP01095804 JAM07911.1 GDIQ01099996 JAL51730.1 GDIQ01115197 JAL36529.1 GDIP01236936 JAI86465.1 GDIQ01251349 JAK00376.1 GDIQ01067606 JAN27131.1 GDIQ01248431 JAK03294.1 GDIP01151597 JAJ71805.1 GDIQ01011177 JAN83560.1 GDIP01010429 JAM93286.1 GDIP01087804 JAM15911.1 JTDY01007475 KOB65170.1
Proteomes
UP000283053
UP000005204
UP000218220
UP000053240
UP000053268
UP000095301
+ More
UP000009192 UP000001819 UP000008744 UP000000803 UP000001292 UP000000304 UP000002282 UP000037069 UP000007798 UP000008711 UP000268350 UP000192221 UP000008792 UP000007801 UP000030765 UP000069272 UP000076408 UP000000673 UP000008820 UP000002320 UP000092553 UP000007266 UP000000311 UP000078200 UP000092444 UP000092445 UP000092462 UP000075885 UP000030742 UP000075880 UP000019118 UP000092443 UP000092460 UP000075883 UP000053825 UP000007062 UP000075840 UP000076502 UP000075920 UP000005203 UP000242457 UP000215335 UP000053105 UP000027135 UP000075900 UP000183832 UP000078542 UP000037510
UP000009192 UP000001819 UP000008744 UP000000803 UP000001292 UP000000304 UP000002282 UP000037069 UP000007798 UP000008711 UP000268350 UP000192221 UP000008792 UP000007801 UP000030765 UP000069272 UP000076408 UP000000673 UP000008820 UP000002320 UP000092553 UP000007266 UP000000311 UP000078200 UP000092444 UP000092445 UP000092462 UP000075885 UP000030742 UP000075880 UP000019118 UP000092443 UP000092460 UP000075883 UP000053825 UP000007062 UP000075840 UP000076502 UP000075920 UP000005203 UP000242457 UP000215335 UP000053105 UP000027135 UP000075900 UP000183832 UP000078542 UP000037510
Interpro
IPR013099
K_chnl_dom
+ More
IPR003092 2pore_dom_K_chnl_TASK
IPR003280 2pore_dom_K_chnl
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR000961 AGC-kinase_C
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR002401 Cyt_P450_E_grp-I
IPR036396 Cyt_P450_sf
IPR017441 Protein_kinase_ATP_BS
IPR003092 2pore_dom_K_chnl_TASK
IPR003280 2pore_dom_K_chnl
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR000961 AGC-kinase_C
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR002401 Cyt_P450_E_grp-I
IPR036396 Cyt_P450_sf
IPR017441 Protein_kinase_ATP_BS
Gene 3D
ProteinModelPortal
A0A2H1VT12
A0A3S2LVT4
H9IUQ3
A0A2W1BYK4
A0A2A4J5D2
A0A0N0PDK3
+ More
A0A194Q8P4 A0A1I8N2A9 A0A0Q9WN14 B4L1K4 Q29HS6 B4GYD9 Q9VYY5 B4IK18 B4R343 B4PYA6 W8AP75 A0A0L0CKA9 B4MSC4 B3NUD1 A0A3B0KEY0 A0A1W4V1W9 B4M355 A0A0K8W0C5 B3MWJ0 A0A0A1WJD1 A0A084VQ45 U5EEQ4 A0A034WRL0 A0A182FMB5 A0A182YH38 W5JI18 Q17DS8 B0XGP9 A0A0M3QZ46 A0A139WIX4 E2B1X3 A0A0K8U275 A0A1Y1KR28 A0A1A9VBQ4 A0A1B0G8B4 A0A1A9Z634 A0A1B0EXA1 A0A182PUM2 U4UHK3 A0A182JKC3 N6UUV7 A0A1A9XXQ9 A0A1B0APF7 A0A182MF40 A0A0L7RDT6 Q7Q822 A7UTI7 A0A182I658 A0A154P4Y7 A0A182W5Z7 A0A088AIB6 A0A2A3ELB4 A0A232F9I3 A0A310S628 A0A0M9A8K7 A0A067QW29 A0A182RVW7 A0A1J1HR05 A0A088ATV4 A0A195CEI0 A0A336KAZ3 A0A0P5I3V3 A0A0P4ZF17 A0A0P5NGR5 A0A0P5APG8 A0A0P5KRQ2 A0A0A9YQN9 A0A0A9XKG5 A0A146M548 A0A0P5SIA8 A0A0A9XF06 A0A0P5KW34 A0A0A9XQD1 A0A0P4YBJ1 A0A0N8B2D2 A0A0P5LTL6 A0A0P5JVA9 A0A0P5JI11 A0A0P6EVF7 A0A0P5T429 A0A0P5VSX9 A0A0P5RLL0 A0A0P5QMF1 A0A0P4XT46 A0A0P5GA70 A0A0P6F080 A0A0P5G0I6 A0A0P5DU13 A0A0P6I569 A0A0P6BV24 A0A0P5WD21 A0A0L7KQ02
A0A194Q8P4 A0A1I8N2A9 A0A0Q9WN14 B4L1K4 Q29HS6 B4GYD9 Q9VYY5 B4IK18 B4R343 B4PYA6 W8AP75 A0A0L0CKA9 B4MSC4 B3NUD1 A0A3B0KEY0 A0A1W4V1W9 B4M355 A0A0K8W0C5 B3MWJ0 A0A0A1WJD1 A0A084VQ45 U5EEQ4 A0A034WRL0 A0A182FMB5 A0A182YH38 W5JI18 Q17DS8 B0XGP9 A0A0M3QZ46 A0A139WIX4 E2B1X3 A0A0K8U275 A0A1Y1KR28 A0A1A9VBQ4 A0A1B0G8B4 A0A1A9Z634 A0A1B0EXA1 A0A182PUM2 U4UHK3 A0A182JKC3 N6UUV7 A0A1A9XXQ9 A0A1B0APF7 A0A182MF40 A0A0L7RDT6 Q7Q822 A7UTI7 A0A182I658 A0A154P4Y7 A0A182W5Z7 A0A088AIB6 A0A2A3ELB4 A0A232F9I3 A0A310S628 A0A0M9A8K7 A0A067QW29 A0A182RVW7 A0A1J1HR05 A0A088ATV4 A0A195CEI0 A0A336KAZ3 A0A0P5I3V3 A0A0P4ZF17 A0A0P5NGR5 A0A0P5APG8 A0A0P5KRQ2 A0A0A9YQN9 A0A0A9XKG5 A0A146M548 A0A0P5SIA8 A0A0A9XF06 A0A0P5KW34 A0A0A9XQD1 A0A0P4YBJ1 A0A0N8B2D2 A0A0P5LTL6 A0A0P5JVA9 A0A0P5JI11 A0A0P6EVF7 A0A0P5T429 A0A0P5VSX9 A0A0P5RLL0 A0A0P5QMF1 A0A0P4XT46 A0A0P5GA70 A0A0P6F080 A0A0P5G0I6 A0A0P5DU13 A0A0P6I569 A0A0P6BV24 A0A0P5WD21 A0A0L7KQ02
PDB
4XDL
E-value=2.78462e-13,
Score=179
Ontologies
GO
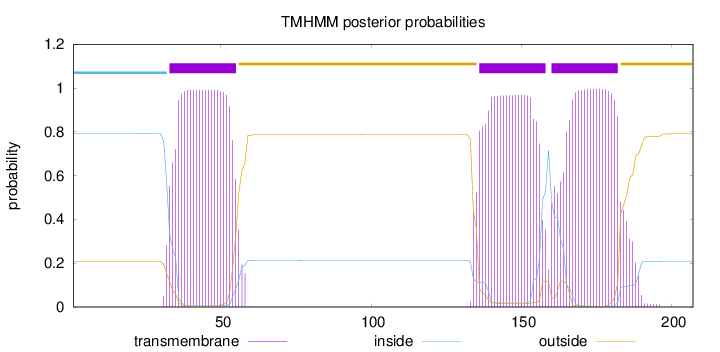
Topology
Length:
207
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
65.67168
Exp number, first 60 AAs:
21.98407
Total prob of N-in:
0.79205
POSSIBLE N-term signal
sequence
inside
1 - 32
TMhelix
33 - 55
outside
56 - 135
TMhelix
136 - 158
inside
159 - 159
TMhelix
160 - 182
outside
183 - 207
Population Genetic Test Statistics
Pi
217.33987
Theta
153.692472
Tajima's D
-0.914025
CLR
1.500358
CSRT
0.15609219539023
Interpretation
Uncertain
