Gene
KWMTBOMO07937
Pre Gene Modal
BGIBMGA000981
Annotation
PREDICTED:_uncharacterized_protein_LOC105841932_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.175
Sequence
CDS
ATGAAGGATTTTATTATTGCACTCCTAGTGTCTATTTGGATTCATCATGGATTTACTCAGATAGCAACATTTGACGTGAGATTAGGGAACTTATACTTAATGCAACGAGGATTAGGGTCGCCATATGAATTAAGAGTAGCAAAGGGACATGAAGCTCTTTTAAAACTGAACAGAGAGTTGGTAAACCAGCAGTCTTGTTCGGTCACAACACCAAAAGGCGTCACGTTTGATGTTCGACAGCCCCCTCAAAACAGATACGAAAGTTGGAGCGATGGATGCGGTGTGAGAGTCCGTAACATAATTAAAGAAGACGAAGGCAGATGGAGGTTAACTGCTACTAGAGGAAATTATTCTATAACTGGATGGTCTGAAGTGCATGTTCAAGATGAAATACCAGAATACCATCCAACACCAATAGCTCTACAAGACGGGGAAAGGCACGCTCATGTTGATCTCACTACGCTCAACAGCGCCTACTGTTTGGTTGAACAGCCCTTTTCTGATATTTCGTTTGTGCCTGGACACTGCAGCGTGACTTTGGATCGGACCACGAGAGCGGTTCAAGGGAACTGGAATGTTGTCCTTGGTCTGCCAGGACAAGTAGCAGAAGTACAAGCCAACACCAAAGTTGAGGTGGAAGGTGAGTTTCGCTATAAATGTACACAGATGCCAAGGGTACGCCAAAGAAAGACTACGAAAGGTCGGGCAGACTTATCAACGTATAAACAAGCTTATGATGATGTAAAAGCGGGATCGTCGTTAAGAACAACGGCAGAAAAACATGGACTGAACCACTGTTCCCTGTTCCGATATGTACATAAACGAGATGCGGCTGGTAACGACGAAAATCAAGAAATGGGATATAAGGCCCACAATCGGGTGTTTACCCGGGAACAAGAGCTTGAGTTATCTAAATATTTAATCAGATGTGCAGATATATATTTTGGCCTAACTAAAAAGGATGTCATGAAATTGGCATACGAACTAACCGTCAAGTACAACTTATCACGACCTCGGACTTGGGATGACAACGGAATGGCTGGCGAAGAATGGTTTCGCATGTTTATGAAAAGAAATTCTGAACTGTCAGTACGTGCAGCACAAGCCACAAGTCTTTCTAGGGCGACTAGCTTTAATAGGAAAAACGTAGATGCTTTCTACGACAATTTGGCTAATGTTATGGACCGCTACAAATTTGAACCACAAAATATCTATAATGTTGATGAGACTGGCATAACTACTGTGCAAAAACCCGACAGAATTATAGCTAGACGTGGTGCTCGTCAAGTAGGCTCAGTAACTTCGGCGGAGAGGGGTGCTTTAGTTACAGTAGCCATTGCGGTCAATGCCATCGGAAATGCTATTCCTCCGTTCTTTGTCTTTCCGCGAGTACGATACCAAGATCACTTTGTCAGGGATGGGCCAATAGGATCTGCTGGGAGTGCAAATCCTTCTGGCTGGATGCAGGATGAATCATTTATGCATTTCCTGGATCACTTCAGGAAACATACGAATGCTTCTCCTTCGCGTAAAATTTTACTTGTGCTTGATAACCACGCGTCACATATACACATTAACGCACTAGACTTTTGCAAGACGAATGGTATTGTAATGCTATCTTTCCCTCCCCACTGCTCACACAAACTCCAACCACTTGATCGATCTGTATTCGGTCCACTAAAAAAAGCTGTAAATTCGACCTGCGATGGATGGATGCGAAGCCACCCCGGAAAGACAATGACAATATACGACATTCCAGGGATTTTAACTACTGCTATGCCGCTTGCTCTTACACAATCCAATATTCAAGCTGGCTTTCGCACAACTGGGATTGTTCCATTTAATCGACATTTGTTTACTGAGCTCGATTTTGCACCTGCCTTTGTGACAGATAGGCCAAATCCATTAGAAGCAGCTGATGGTCCTATTCAGAATATTAATACACTTGAGGATAAAACGCCACCAACATCACCTTCCATTTTAACCCTCGAACCACAAGAAGAGATGGAGGAGCTTTCTAATGAAGCTCTTCTTGTACAACAACCGACTGAAAATATGCCACAATGTTCACAAGTACTTGATAAAGAACAGGAAAACATTATTGCAAATCCAAAACTTTTGATTATATTACCCGGACATGAAAGTAGTAGTAGCGCTCAGCAAATTTCACCCAAACCCTCTAAAAGTTCTCAATCCATTCCTGTGTTAACAACTCAAACGGGAAGTATCATGACTTTGACTACACCATCTTTATTTTCGCCAGAAGTAATACGACCACTACCAAAAGCTCCTCCAAGGAAATTAACAAATAGGGGCCGAAAAACCAGGAAATCTACTATCTACACAGACACACCTGAAAAAGAAGAGATAAGAAGAGAATATGAAAACAGATTGAAACGAACCAAAGCTAAACAAGTTAAGAAAAGATTAGATGGGGGAAAGACTAAAACAAATGCTAGAAGCAGGGGAAAAATTAAGATGCACCAAGAGTCATCATCATCAGAAGAAGAGGAGTGCTACTGTGTTGTTTGTATGTCAGCATATTCAGAAAGCAGACCCAGGGAAAAATGGATACAATGCACAGTATGTAAAATGTGGGCGCATGAAGAGTGCACACAAGTTGAAAGTTTGGACACGGGATACGTACAAGATACGAATGCTAAGAAGTTACATTTGTACTGCAACATATTGCATACACAGAAGAATATCACATTTTGTCGTTTCCAAAAGATAACGGAACATGTTGGTTATAATATTATGAACGGCTTGAGCGATGGTGCGCACAGTTATTACGGGGACGGTTTCGAGATGCGACACTGTGGTATGACGGTGGAAAATCCAACTGAACAAGTGTTCGGTACGTGGCGGTGTACTGTGGGAGTTCAGGAAAGGGTCGGCACTCAAATACACCAGAGGACACCGATGCAAGCGTTGATTAGGGTTTCCCCATACAATTTAGGTTCAAGAATCATGAATGAAGTTAATGAAGACAGCATTGCGAGTAGAACGATCTTTGTACAAAGGGATATGGGTTTCACAATAACGTGCAGAGCTGAGGTTTCATTGAGCTATTGTTGGTTTCAACATCCGAACGGAACCCAGTACACTCCTGTGCCACGGAGCGATGATTCCGATGACAGTTTATTTTGGTATGCCGGTGAAAGTCTTCAAACTGGCGATTGTGGTATATCGTTTTCTTACGCTACGGATGAAGACTCCGGAGAATGGACTTGTCATATGGGTCCAAGAAGGCAAATGGGAGTCGAACTTACCGATAAATTAGTAGTTCGTGTGACTGGTCCGTTAGCGGCTAGTCAAAAGGAGATCCCTACGGTGATTGATGGGAGTGCTACATTGTTTTGTAAAACGTCGAATGGGAACAGACCGTTGGAATACTGTCGGTTCTTGTCCCCAAAATTTGTTGGCATTAGCATTGATCCGTCTGTTACAAAGGAAAACGCAATACTGGGCAGATACTTTTTTACAACCGGCAGAGATTTAGATTATGGTGATTGTTCTTTGACGATTATATCGGTAACCAATGATGACTTAGGAGAGTGGACATGCGCTGCTCTACTACACGACGAGGCCGCGGAGAGTAGAGACATCATGACACTCTACGTCGAACAAAGAAGATTGGAATGA
Protein
MKDFIIALLVSIWIHHGFTQIATFDVRLGNLYLMQRGLGSPYELRVAKGHEALLKLNRELVNQQSCSVTTPKGVTFDVRQPPQNRYESWSDGCGVRVRNIIKEDEGRWRLTATRGNYSITGWSEVHVQDEIPEYHPTPIALQDGERHAHVDLTTLNSAYCLVEQPFSDISFVPGHCSVTLDRTTRAVQGNWNVVLGLPGQVAEVQANTKVEVEGEFRYKCTQMPRVRQRKTTKGRADLSTYKQAYDDVKAGSSLRTTAEKHGLNHCSLFRYVHKRDAAGNDENQEMGYKAHNRVFTREQELELSKYLIRCADIYFGLTKKDVMKLAYELTVKYNLSRPRTWDDNGMAGEEWFRMFMKRNSELSVRAAQATSLSRATSFNRKNVDAFYDNLANVMDRYKFEPQNIYNVDETGITTVQKPDRIIARRGARQVGSVTSAERGALVTVAIAVNAIGNAIPPFFVFPRVRYQDHFVRDGPIGSAGSANPSGWMQDESFMHFLDHFRKHTNASPSRKILLVLDNHASHIHINALDFCKTNGIVMLSFPPHCSHKLQPLDRSVFGPLKKAVNSTCDGWMRSHPGKTMTIYDIPGILTTAMPLALTQSNIQAGFRTTGIVPFNRHLFTELDFAPAFVTDRPNPLEAADGPIQNINTLEDKTPPTSPSILTLEPQEEMEELSNEALLVQQPTENMPQCSQVLDKEQENIIANPKLLIILPGHESSSSAQQISPKPSKSSQSIPVLTTQTGSIMTLTTPSLFSPEVIRPLPKAPPRKLTNRGRKTRKSTIYTDTPEKEEIRREYENRLKRTKAKQVKKRLDGGKTKTNARSRGKIKMHQESSSSEEEECYCVVCMSAYSESRPREKWIQCTVCKMWAHEECTQVESLDTGYVQDTNAKKLHLYCNILHTQKNITFCRFQKITEHVGYNIMNGLSDGAHSYYGDGFEMRHCGMTVENPTEQVFGTWRCTVGVQERVGTQIHQRTPMQALIRVSPYNLGSRIMNEVNEDSIASRTIFVQRDMGFTITCRAEVSLSYCWFQHPNGTQYTPVPRSDDSDDSLFWYAGESLQTGDCGISFSYATDEDSGEWTCHMGPRRQMGVELTDKLVVRVTGPLAASQKEIPTVIDGSATLFCKTSNGNRPLEYCRFLSPKFVGISIDPSVTKENAILGRYFFTTGRDLDYGDCSLTIISVTNDDLGEWTCAALLHDEAAESRDIMTLYVEQRRLE
Summary
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
Pfam
PF03184 DDE_1
Interpro
Gene 3D
ProteinModelPortal
PDB
1F97
E-value=0.0120724,
Score=96
Ontologies
GO
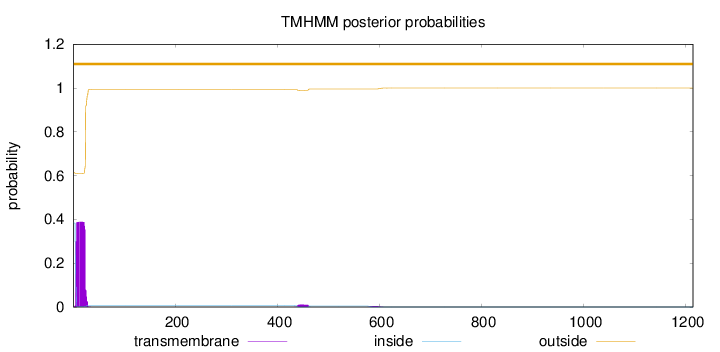
Topology
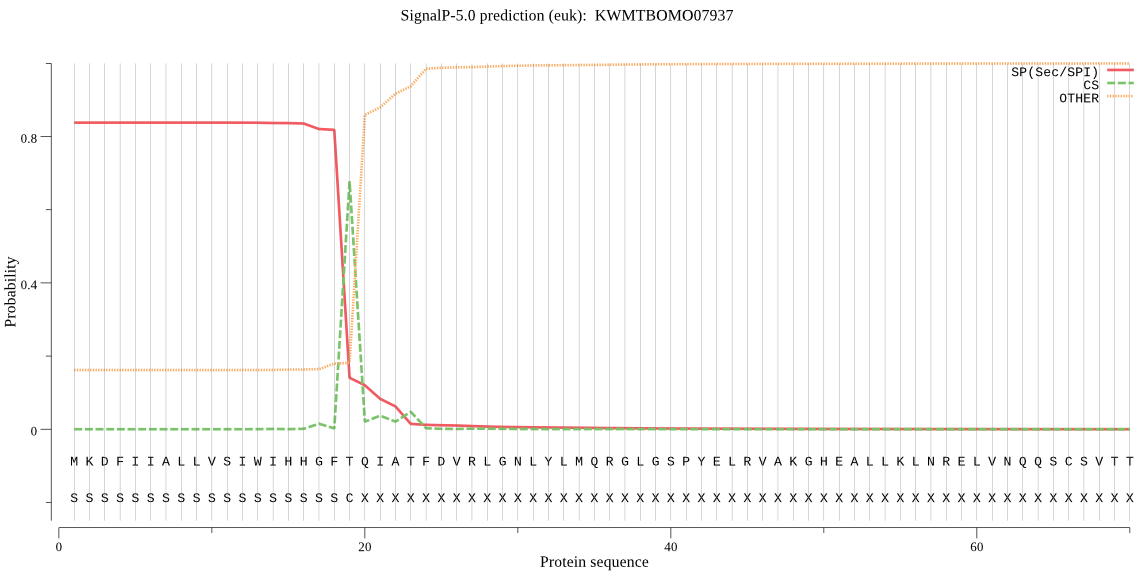
SignalP
Position: 1 - 19,
Likelihood: 0.837019
Length:
1214
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.13118999999996
Exp number, first 60 AAs:
7.80991
Total prob of N-in:
0.38570
outside
1 - 1214
Population Genetic Test Statistics
Pi
167.912258
Theta
151.769056
Tajima's D
-1.079177
CLR
50.186271
CSRT
0.124743762811859
Interpretation
Uncertain
