Gene
KWMTBOMO07936
Pre Gene Modal
BGIBMGA000980
Annotation
PREDICTED:_TIP41-like_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.92
Sequence
CDS
ATGTTTCGACCAGAAGAAACAAATGTTGATGCCGGTCGTCATACGTCTAATTCGCAGTCCATTGAATTTGGTCCTTGGCATATTTCGTATGATGTCAGCCGTATACTACCCAGTATGTGTGCCACTAAGGTGGTTTGCGAGAGAGATGACGACCAATTTTGTCAGTTCTGCACTTACTCTAAAGAGCTGAAGATTCCACATTTCCCTGATATGGTATTTCCAAAAAACAGATTAACATTAGCACATAAAAGCGGTGCTAGTATAAACTTTAATCCTCTGGATGCTCTTAAAAGGGTGGCTAGCACAGTGGAGCCAGTTGAAGTGTCCTGTTCTGAAGTGTGGATGCAAGCACGACCATATGCAGAAAAGTTGAAGAAATCATTTGATTGGACTTTTTGTACAGATTACAAAGGATCTATCAGTGATAATATTACAGTTTGGGAAACTGAAGAGTCTATTGATTTTGAACTGTTAAAACAGAAAAACCAGATACTCTTCTACCATGATTTGACTTTATTTGAAGATGAGCTGCATGATCATGGTGTATCAAAGCTCTCAGTGAAAATTAGAGTTATGCCATCCTATTGGTTTGTTTTGTTGAGATATTTTTTGAGAGTGGATGATGTCCTAGTCAGATCAAATGAAACTAGAATGTTTCATATGCTTAACACAGATTACATACTAAGAGAATATACTTCTAAAGAATCGAGCGCAAAGGATTTGAATATGCCCACGTCCCACATGAAGGAAGCAGATGACGTCATTCCATTTTTACCTGTCAAGGAGAAAATTACACATAAGCTTCATTTTAATTGTAGTTCCTAG
Protein
MFRPEETNVDAGRHTSNSQSIEFGPWHISYDVSRILPSMCATKVVCERDDDQFCQFCTYSKELKIPHFPDMVFPKNRLTLAHKSGASINFNPLDALKRVASTVEPVEVSCSEVWMQARPYAEKLKKSFDWTFCTDYKGSISDNITVWETEESIDFELLKQKNQILFYHDLTLFEDELHDHGVSKLSVKIRVMPSYWFVLLRYFLRVDDVLVRSNETRMFHMLNTDYILREYTSKESSAKDLNMPTSHMKEADDVIPFLPVKEKITHKLHFNCSS
Summary
Uniprot
A0A1E1WW32
A0A2A4J4W7
A0A3S2NE33
A0A2H1W9E6
A0A194Q8P9
A0A2A4J620
+ More
A0A212F469 A0A0N1IIN3 A0A0L7KSV4 A0A2J7Q4Z5 A0A1B6FYG0 A0A1W4XU35 D1ZZJ5 A0A2P8XHY0 A0A154P550 V5I6X7 A0A0C9Q9S8 E2BGM5 A0A0K8TQH5 N6U713 A0A026W3N4 B3MQI6 E9ILR5 A0A1Q3G2N8 A0A1W4US88 A0A1W4UDS6 Q16M33 A0A3L8E2V7 B4M9Y1 J3JW78 A0A1B6C2K4 A0A1A9XXT8 A0A0M9A791 A0A1S4FWM6 B4PYX2 A0A1B0APQ4 A0A0L7RD57 K7IYT0 B4JK97 A0A023ENE3 A0A088AIL1 A0A195F8Y3 A0A1A9VBL0 A0A1B0G8D7 A0A1A9Z5V7 A0A034WKE1 W8BFH2 A0A0K8V082 A0A1I8MGR7 A0A195CI89 A0A1B6H8D7 A0A182JTU2 A0A1B6LF12 B4L7X4 A0A1B6KCS4 A0A0Q9XG04 F4WF53 B3NWG5 A0A182QKB3 B4NE81 A0A182V5Y1 A0A1A9W223 A0A182XAI8 A0A182U416 E0VH67 A0A2A3EJS6 A0A182NRA1 A0A182LEC0 Q5TTX3 A0A182HSC8 A0A182RQL0 A0A158NP56 Q29GW1 A0A1I8PHJ8 A0A0R3P6I6 A0A182Y6Q4 A0A182VZY9 B4GV23 E2B1X4 A0A182SAW3 A0A195DZ03 A0A182P3V2 A0A0M3QWM9 A0A151WXY4 A0A182GEQ4 Q9W5W6 A0A3B0K4V1 W5J762 A0A232EYE1 A0A182FAH4 A0A195AY39 A0A1Y1LPK6 A0A182J828 A0A1L8DWS7 A0A1S3DHQ9 A0A1L8DWB3 A0A131YC75 A0A3B0J8G9
A0A212F469 A0A0N1IIN3 A0A0L7KSV4 A0A2J7Q4Z5 A0A1B6FYG0 A0A1W4XU35 D1ZZJ5 A0A2P8XHY0 A0A154P550 V5I6X7 A0A0C9Q9S8 E2BGM5 A0A0K8TQH5 N6U713 A0A026W3N4 B3MQI6 E9ILR5 A0A1Q3G2N8 A0A1W4US88 A0A1W4UDS6 Q16M33 A0A3L8E2V7 B4M9Y1 J3JW78 A0A1B6C2K4 A0A1A9XXT8 A0A0M9A791 A0A1S4FWM6 B4PYX2 A0A1B0APQ4 A0A0L7RD57 K7IYT0 B4JK97 A0A023ENE3 A0A088AIL1 A0A195F8Y3 A0A1A9VBL0 A0A1B0G8D7 A0A1A9Z5V7 A0A034WKE1 W8BFH2 A0A0K8V082 A0A1I8MGR7 A0A195CI89 A0A1B6H8D7 A0A182JTU2 A0A1B6LF12 B4L7X4 A0A1B6KCS4 A0A0Q9XG04 F4WF53 B3NWG5 A0A182QKB3 B4NE81 A0A182V5Y1 A0A1A9W223 A0A182XAI8 A0A182U416 E0VH67 A0A2A3EJS6 A0A182NRA1 A0A182LEC0 Q5TTX3 A0A182HSC8 A0A182RQL0 A0A158NP56 Q29GW1 A0A1I8PHJ8 A0A0R3P6I6 A0A182Y6Q4 A0A182VZY9 B4GV23 E2B1X4 A0A182SAW3 A0A195DZ03 A0A182P3V2 A0A0M3QWM9 A0A151WXY4 A0A182GEQ4 Q9W5W6 A0A3B0K4V1 W5J762 A0A232EYE1 A0A182FAH4 A0A195AY39 A0A1Y1LPK6 A0A182J828 A0A1L8DWS7 A0A1S3DHQ9 A0A1L8DWB3 A0A131YC75 A0A3B0J8G9
Pubmed
26354079
22118469
26227816
18362917
19820115
29403074
+ More
20798317 26369729 23537049 24508170 17994087 21282665 17510324 30249741 22516182 17550304 20075255 24945155 25348373 24495485 25315136 21719571 20566863 20966253 12364791 14747013 17210077 21347285 15632085 25244985 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 28648823 28004739 26830274
20798317 26369729 23537049 24508170 17994087 21282665 17510324 30249741 22516182 17550304 20075255 24945155 25348373 24495485 25315136 21719571 20566863 20966253 12364791 14747013 17210077 21347285 15632085 25244985 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 28648823 28004739 26830274
EMBL
GDQN01000073
JAT90981.1
NWSH01002968
PCG67187.1
RSAL01000173
RVE45079.1
+ More
ODYU01007173 SOQ49719.1 KQ459299 KPJ01912.1 PCG67186.1 AGBW02010426 OWR48536.1 KQ460202 KPJ17178.1 JTDY01006305 KOB66119.1 NEVH01018372 PNF23659.1 GECZ01014560 JAS55209.1 KQ971338 EFA01841.1 PYGN01002069 PSN31607.1 KQ434820 KZC06983.1 GALX01007415 JAB61051.1 GBYB01011127 GBYB01011128 JAG80894.1 JAG80895.1 GL448193 EFN85153.1 GDAI01000986 JAI16617.1 APGK01040221 APGK01040222 KB740978 ENN76436.1 KK107468 EZA50221.1 CH902621 EDV44612.1 GL764129 EFZ18460.1 GFDL01000982 JAV34063.1 CH477882 EAT35384.1 QOIP01000001 RLU27071.1 CH940655 EDW66040.1 BT127496 AEE62458.1 GEDC01029541 JAS07757.1 KQ435720 KOX78675.1 CM000162 EDX02050.1 JXJN01001542 KQ414614 KOC68897.1 CH916370 EDV99999.1 GAPW01003168 JAC10430.1 KQ981727 KYN36841.1 CCAG010000247 GAKP01002896 JAC56056.1 GAMC01010862 JAB95693.1 GDHF01019987 JAI32327.1 KQ977791 KYM99793.1 GECU01036748 JAS70958.1 GEBQ01017705 JAT22272.1 CH933814 EDW05549.1 GEBQ01030741 JAT09236.1 KRG07446.1 GL888115 EGI67104.1 CH954180 EDV46785.1 AXCN02001467 CH964239 EDW82050.1 DS235161 EEB12723.1 KZ288222 PBC32063.1 AAAB01008859 EAL40866.3 APCN01000261 ADTU01022241 CH379064 EAL31998.2 KRT06758.1 CH479192 EDW26560.1 GL444981 EFN60330.1 KQ980066 KYN17952.1 CP012525 ALC44429.1 KQ982668 KYQ52551.1 JXUM01162811 KQ574605 KXJ67859.1 AE014298 AY058495 AAF45376.2 AAL13724.1 OUUW01000003 SPP78498.1 ADMH02002037 ETN59816.1 NNAY01001634 OXU23363.1 KQ976703 KYM77163.1 GEZM01051682 JAV74748.1 GFDF01003359 JAV10725.1 GFDF01003358 JAV10726.1 GEDV01012462 JAP76095.1 SPP78497.1
ODYU01007173 SOQ49719.1 KQ459299 KPJ01912.1 PCG67186.1 AGBW02010426 OWR48536.1 KQ460202 KPJ17178.1 JTDY01006305 KOB66119.1 NEVH01018372 PNF23659.1 GECZ01014560 JAS55209.1 KQ971338 EFA01841.1 PYGN01002069 PSN31607.1 KQ434820 KZC06983.1 GALX01007415 JAB61051.1 GBYB01011127 GBYB01011128 JAG80894.1 JAG80895.1 GL448193 EFN85153.1 GDAI01000986 JAI16617.1 APGK01040221 APGK01040222 KB740978 ENN76436.1 KK107468 EZA50221.1 CH902621 EDV44612.1 GL764129 EFZ18460.1 GFDL01000982 JAV34063.1 CH477882 EAT35384.1 QOIP01000001 RLU27071.1 CH940655 EDW66040.1 BT127496 AEE62458.1 GEDC01029541 JAS07757.1 KQ435720 KOX78675.1 CM000162 EDX02050.1 JXJN01001542 KQ414614 KOC68897.1 CH916370 EDV99999.1 GAPW01003168 JAC10430.1 KQ981727 KYN36841.1 CCAG010000247 GAKP01002896 JAC56056.1 GAMC01010862 JAB95693.1 GDHF01019987 JAI32327.1 KQ977791 KYM99793.1 GECU01036748 JAS70958.1 GEBQ01017705 JAT22272.1 CH933814 EDW05549.1 GEBQ01030741 JAT09236.1 KRG07446.1 GL888115 EGI67104.1 CH954180 EDV46785.1 AXCN02001467 CH964239 EDW82050.1 DS235161 EEB12723.1 KZ288222 PBC32063.1 AAAB01008859 EAL40866.3 APCN01000261 ADTU01022241 CH379064 EAL31998.2 KRT06758.1 CH479192 EDW26560.1 GL444981 EFN60330.1 KQ980066 KYN17952.1 CP012525 ALC44429.1 KQ982668 KYQ52551.1 JXUM01162811 KQ574605 KXJ67859.1 AE014298 AY058495 AAF45376.2 AAL13724.1 OUUW01000003 SPP78498.1 ADMH02002037 ETN59816.1 NNAY01001634 OXU23363.1 KQ976703 KYM77163.1 GEZM01051682 JAV74748.1 GFDF01003359 JAV10725.1 GFDF01003358 JAV10726.1 GEDV01012462 JAP76095.1 SPP78497.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000235965 UP000192223 UP000007266 UP000245037 UP000076502 UP000008237 UP000019118 UP000053097 UP000007801 UP000192221 UP000008820 UP000279307 UP000008792 UP000092443 UP000053105 UP000002282 UP000092460 UP000053825 UP000002358 UP000001070 UP000005203 UP000078541 UP000078200 UP000092444 UP000092445 UP000095301 UP000078542 UP000075881 UP000009192 UP000007755 UP000008711 UP000075886 UP000007798 UP000075903 UP000091820 UP000076407 UP000075902 UP000009046 UP000242457 UP000075884 UP000075882 UP000007062 UP000075840 UP000075900 UP000005205 UP000001819 UP000095300 UP000076408 UP000075920 UP000008744 UP000000311 UP000075901 UP000078492 UP000075885 UP000092553 UP000075809 UP000069940 UP000249989 UP000000803 UP000268350 UP000000673 UP000215335 UP000069272 UP000078540 UP000075880 UP000079169
UP000235965 UP000192223 UP000007266 UP000245037 UP000076502 UP000008237 UP000019118 UP000053097 UP000007801 UP000192221 UP000008820 UP000279307 UP000008792 UP000092443 UP000053105 UP000002282 UP000092460 UP000053825 UP000002358 UP000001070 UP000005203 UP000078541 UP000078200 UP000092444 UP000092445 UP000095301 UP000078542 UP000075881 UP000009192 UP000007755 UP000008711 UP000075886 UP000007798 UP000075903 UP000091820 UP000076407 UP000075902 UP000009046 UP000242457 UP000075884 UP000075882 UP000007062 UP000075840 UP000075900 UP000005205 UP000001819 UP000095300 UP000076408 UP000075920 UP000008744 UP000000311 UP000075901 UP000078492 UP000075885 UP000092553 UP000075809 UP000069940 UP000249989 UP000000803 UP000268350 UP000000673 UP000215335 UP000069272 UP000078540 UP000075880 UP000079169
PRIDE
Pfam
PF04176 TIP41
SUPFAM
SSF48726
SSF48726
ProteinModelPortal
A0A1E1WW32
A0A2A4J4W7
A0A3S2NE33
A0A2H1W9E6
A0A194Q8P9
A0A2A4J620
+ More
A0A212F469 A0A0N1IIN3 A0A0L7KSV4 A0A2J7Q4Z5 A0A1B6FYG0 A0A1W4XU35 D1ZZJ5 A0A2P8XHY0 A0A154P550 V5I6X7 A0A0C9Q9S8 E2BGM5 A0A0K8TQH5 N6U713 A0A026W3N4 B3MQI6 E9ILR5 A0A1Q3G2N8 A0A1W4US88 A0A1W4UDS6 Q16M33 A0A3L8E2V7 B4M9Y1 J3JW78 A0A1B6C2K4 A0A1A9XXT8 A0A0M9A791 A0A1S4FWM6 B4PYX2 A0A1B0APQ4 A0A0L7RD57 K7IYT0 B4JK97 A0A023ENE3 A0A088AIL1 A0A195F8Y3 A0A1A9VBL0 A0A1B0G8D7 A0A1A9Z5V7 A0A034WKE1 W8BFH2 A0A0K8V082 A0A1I8MGR7 A0A195CI89 A0A1B6H8D7 A0A182JTU2 A0A1B6LF12 B4L7X4 A0A1B6KCS4 A0A0Q9XG04 F4WF53 B3NWG5 A0A182QKB3 B4NE81 A0A182V5Y1 A0A1A9W223 A0A182XAI8 A0A182U416 E0VH67 A0A2A3EJS6 A0A182NRA1 A0A182LEC0 Q5TTX3 A0A182HSC8 A0A182RQL0 A0A158NP56 Q29GW1 A0A1I8PHJ8 A0A0R3P6I6 A0A182Y6Q4 A0A182VZY9 B4GV23 E2B1X4 A0A182SAW3 A0A195DZ03 A0A182P3V2 A0A0M3QWM9 A0A151WXY4 A0A182GEQ4 Q9W5W6 A0A3B0K4V1 W5J762 A0A232EYE1 A0A182FAH4 A0A195AY39 A0A1Y1LPK6 A0A182J828 A0A1L8DWS7 A0A1S3DHQ9 A0A1L8DWB3 A0A131YC75 A0A3B0J8G9
A0A212F469 A0A0N1IIN3 A0A0L7KSV4 A0A2J7Q4Z5 A0A1B6FYG0 A0A1W4XU35 D1ZZJ5 A0A2P8XHY0 A0A154P550 V5I6X7 A0A0C9Q9S8 E2BGM5 A0A0K8TQH5 N6U713 A0A026W3N4 B3MQI6 E9ILR5 A0A1Q3G2N8 A0A1W4US88 A0A1W4UDS6 Q16M33 A0A3L8E2V7 B4M9Y1 J3JW78 A0A1B6C2K4 A0A1A9XXT8 A0A0M9A791 A0A1S4FWM6 B4PYX2 A0A1B0APQ4 A0A0L7RD57 K7IYT0 B4JK97 A0A023ENE3 A0A088AIL1 A0A195F8Y3 A0A1A9VBL0 A0A1B0G8D7 A0A1A9Z5V7 A0A034WKE1 W8BFH2 A0A0K8V082 A0A1I8MGR7 A0A195CI89 A0A1B6H8D7 A0A182JTU2 A0A1B6LF12 B4L7X4 A0A1B6KCS4 A0A0Q9XG04 F4WF53 B3NWG5 A0A182QKB3 B4NE81 A0A182V5Y1 A0A1A9W223 A0A182XAI8 A0A182U416 E0VH67 A0A2A3EJS6 A0A182NRA1 A0A182LEC0 Q5TTX3 A0A182HSC8 A0A182RQL0 A0A158NP56 Q29GW1 A0A1I8PHJ8 A0A0R3P6I6 A0A182Y6Q4 A0A182VZY9 B4GV23 E2B1X4 A0A182SAW3 A0A195DZ03 A0A182P3V2 A0A0M3QWM9 A0A151WXY4 A0A182GEQ4 Q9W5W6 A0A3B0K4V1 W5J762 A0A232EYE1 A0A182FAH4 A0A195AY39 A0A1Y1LPK6 A0A182J828 A0A1L8DWS7 A0A1S3DHQ9 A0A1L8DWB3 A0A131YC75 A0A3B0J8G9
PDB
5W0W
E-value=2.08644e-44,
Score=449
Ontologies
PANTHER
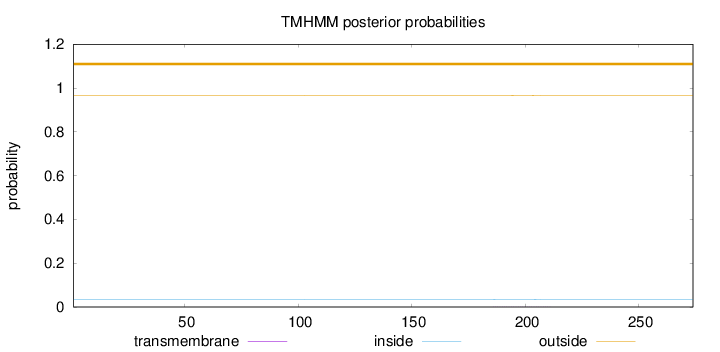
Topology
Length:
274
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01149
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03396
outside
1 - 274
Population Genetic Test Statistics
Pi
174.680939
Theta
184.086946
Tajima's D
-0.296229
CLR
0.975731
CSRT
0.283485825708715
Interpretation
Uncertain
