Gene
KWMTBOMO07934
Annotation
PREDICTED:_gamma-tubulin_complex_component_2-like_isoform_X2_[Bombyx_mori]
Full name
Gamma-tubulin complex component
+ More
Gamma-tubulin complex component 2 homolog
Gamma-tubulin complex component 2 homolog
Alternative Name
Gamma-ring complex protein 84 kDa
Location in the cell
Extracellular Reliability : 2.766
Sequence
CDS
ATGAGTTTTACGAATAAATTTCAGGTGCACAACGTTGATGAAGTGTTGGAGAGGCACAATGACTTCCTCGAATCCTGCCTCGGGGACTGCATGCTCACTAATCCTCAACTGTTGAAGGCTGTCACCACGCTCTGTCTCGTTTGCGTCCAGTTCTGCAGTTTCATACAGGAGGCAGGTTGCGGGACCACTACGAGCAGCTCGGCGGACTCATTCTCTCGTTCTGTGTCGCGGTACGGGCTTCGATTCACGGCGGCACTTCTCTCCGTGCTCGCTATAATCGATCGTAATGCACGCGACAACAACACGAACAAACTCCTCAACATATCCGCCAGATTGAATTTCAACACTTATTACGCGAAGCAACTTGAAAAGTTTTGCTCTGATGACAAACTTCTGGACTGTGAGAAGAAAACGCCGTCCTCCTAG
Protein
MSFTNKFQVHNVDEVLERHNDFLESCLGDCMLTNPQLLKAVTTLCLVCVQFCSFIQEAGCGTTTSSSADSFSRSVSRYGLRFTAALLSVLAIIDRNARDNNTNKLLNISARLNFNTYYAKQLEKFCSDDKLLDCEKKTPSS
Summary
Description
Gamma-tubulin complex is necessary for microtubule nucleation at the centrosome.
Subunit
Gamma-tubulin small complex (Gamma TuSC) is a heterotetrameric complex which contains two molecules of gamma-tubulin, and one molecule each of Dgrip84 and Dgrip91. The gamma-tubulin in this complex binds preferentially to GDP over GTP.
Similarity
Belongs to the TUBGCP family.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
Cytoskeleton
Direct protein sequencing
Microtubule
Phosphoprotein
Reference proteome
Feature
chain Gamma-tubulin complex component 2 homolog
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2H1VKZ0
A0A2A4J4X6
A0A2A4J696
A0A212EZL8
A0A2W1BUG7
A0A194QA90
+ More
A0A3S2NUS6 A0A0L7KTL8 A0A2J7PCE5 A0A2J7PCF9 A0A2P8Y7R0 A0A187GBR9 A0A2J7PCE4 A0A0A1XS79 A0A0A1WM46 A0A067QYV0 A0A0A1WF42 W8B8N2 A0A034W254 A0A0K8VC34 A0A0K8U1N3 A0A0K8UQ43 A0A0K8V489 A0A1W4VVE2 A0A1A9VWY4 A0A1A9Z3W0 A0A1I8NA16 A0A2M4BEW0 A0A1A9W9K4 A0A0P4WIJ5 A0A0R3P5S0 A0A1I8PP14 A0A1Q3G1T4 A0A2M4AHX1 A0A2M4AHX5 A0A0Q5T4B1 A0A2M3Z798 A0A2M4BDP6 A0A2M3Z7H8 A0A2M4BDN9 A0A0Q9WDN2 A0A0Q9XF36 Q16HU3 W5JU97 W8BFW3 A0A3B0KWP9 A0A3B0KMD8 A0A0A1WYP6 A0A0R1EAT5 A0A1A9WFR2 A0A0K8W6D2 A0A034W3R9 A0A1B0G596 A0A1W4VHA8 E1JJQ3 A0A0L0BRN3 Q9XYP7-2 X2JG40 A0A1I8NA10 A0A1I8NA08 A0A2B4RSY1 B4NCC7 A0A0T6B308 D2A387 A0A3F2YSR6 Q7Q703 A0A1Q3G1T0 A0A3B0J9L1 A0A3R7M2R8 A0A1Y1NJV3 A0A084WKN6 Q29G36 B4GW52 A0A182PHG5 A0A1B6FY55 A0A182M1W1 A0A336MC89 A0A2R9BTL5 A0A182HDF5 Q16HU4 A0A1B6IKE5 B3NVP5 G5AVB2 A0A091E667 B4PZH1 A0A182Y143 A0A026WSN5 A0A232EUY2 A0A151IHY4 K7IYU5 A0A1J1HG27 F4WNM3 A0A0M3QTM0 A0A195B7L9 A0A158NUW2 A0A195FV78
A0A3S2NUS6 A0A0L7KTL8 A0A2J7PCE5 A0A2J7PCF9 A0A2P8Y7R0 A0A187GBR9 A0A2J7PCE4 A0A0A1XS79 A0A0A1WM46 A0A067QYV0 A0A0A1WF42 W8B8N2 A0A034W254 A0A0K8VC34 A0A0K8U1N3 A0A0K8UQ43 A0A0K8V489 A0A1W4VVE2 A0A1A9VWY4 A0A1A9Z3W0 A0A1I8NA16 A0A2M4BEW0 A0A1A9W9K4 A0A0P4WIJ5 A0A0R3P5S0 A0A1I8PP14 A0A1Q3G1T4 A0A2M4AHX1 A0A2M4AHX5 A0A0Q5T4B1 A0A2M3Z798 A0A2M4BDP6 A0A2M3Z7H8 A0A2M4BDN9 A0A0Q9WDN2 A0A0Q9XF36 Q16HU3 W5JU97 W8BFW3 A0A3B0KWP9 A0A3B0KMD8 A0A0A1WYP6 A0A0R1EAT5 A0A1A9WFR2 A0A0K8W6D2 A0A034W3R9 A0A1B0G596 A0A1W4VHA8 E1JJQ3 A0A0L0BRN3 Q9XYP7-2 X2JG40 A0A1I8NA10 A0A1I8NA08 A0A2B4RSY1 B4NCC7 A0A0T6B308 D2A387 A0A3F2YSR6 Q7Q703 A0A1Q3G1T0 A0A3B0J9L1 A0A3R7M2R8 A0A1Y1NJV3 A0A084WKN6 Q29G36 B4GW52 A0A182PHG5 A0A1B6FY55 A0A182M1W1 A0A336MC89 A0A2R9BTL5 A0A182HDF5 Q16HU4 A0A1B6IKE5 B3NVP5 G5AVB2 A0A091E667 B4PZH1 A0A182Y143 A0A026WSN5 A0A232EUY2 A0A151IHY4 K7IYU5 A0A1J1HG27 F4WNM3 A0A0M3QTM0 A0A195B7L9 A0A158NUW2 A0A195FV78
Pubmed
22118469
28756777
26354079
26227816
29403074
25830018
+ More
24845553 24495485 25348373 25315136 15632085 17994087 17510324 20920257 23761445 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 10037793 12537569 17372656 18362917 19820115 12364791 28004739 24438588 22722832 26483478 21993625 25244985 24508170 28648823 20075255 21719571 21347285
24845553 24495485 25348373 25315136 15632085 17994087 17510324 20920257 23761445 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 10037793 12537569 17372656 18362917 19820115 12364791 28004739 24438588 22722832 26483478 21993625 25244985 24508170 28648823 20075255 21719571 21347285
EMBL
ODYU01003123
SOQ41485.1
NWSH01002968
PCG67197.1
PCG67198.1
AGBW02011255
+ More
OWR46930.1 KZ149944 PZC76847.1 KQ459299 KPJ01915.1 RSAL01000173 RVE45081.1 JTDY01006012 KOB66374.1 NEVH01027059 PNF14009.1 PNF14012.1 PYGN01000836 PSN40194.1 KR733678 AKS04550.1 PNF14010.1 GBXI01000537 JAD13755.1 GBXI01014819 JAC99472.1 KK852868 KDR14696.1 GBXI01016770 JAC97521.1 GAMC01008935 JAB97620.1 GAKP01010198 JAC48754.1 GDHF01015917 JAI36397.1 GDHF01031692 JAI20622.1 GDHF01023613 JAI28701.1 GDHF01018633 JAI33681.1 GGFJ01002449 MBW51590.1 GDRN01030756 JAI67539.1 CH379064 KRT07119.1 GFDL01001285 JAV33760.1 GGFK01007050 MBW40371.1 GGFK01007059 MBW40380.1 CH954180 KQS30093.1 GGFM01003630 MBW24381.1 GGFJ01002024 MBW51165.1 GGFM01003710 MBW24461.1 GGFJ01002025 MBW51166.1 CH940655 KRF82554.1 CH933812 KRG07147.1 CH478141 EAT33821.1 ADMH02000350 ETN66868.1 GAMC01008933 JAB97622.1 OUUW01000031 SPP89791.1 SPP89790.1 GBXI01010759 JAD03533.1 CM000162 KRK06576.1 GDHF01005874 JAI46440.1 GAKP01010197 JAC48755.1 CCAG010022616 AE014298 ACZ95332.1 JRES01001467 KNC22653.1 AF118379 AY061148 AHN59914.1 LSMT01000338 PFX19929.1 CH964239 EDW82486.1 LJIG01016094 KRT81633.1 KQ971338 EFA02278.2 APCN01004264 AAAB01008960 EAA11766.4 GFDL01001281 JAV33764.1 OUUW01000002 SPP77053.1 QCYY01002451 ROT70212.1 GEZM01001085 GEZM01001084 JAV98171.1 ATLV01024126 KE525349 KFB50780.1 EAL32274.1 CH479193 EDW26897.1 GECZ01014644 JAS55125.1 AXCM01006421 UFQT01000888 SSX27846.1 AJFE02055016 AJFE02055017 AJFE02055018 AJFE02055019 JXUM01034315 JXUM01034316 JXUM01034317 KQ561019 KXJ80013.1 EAT33820.1 GECU01020320 JAS87386.1 EDV46710.2 JH167069 EHB00973.1 KN120575 KFO38223.1 EDX02127.1 KK107109 EZA59060.1 NNAY01002072 OXU22162.1 KQ977545 KYN02030.1 CVRI01000002 CRK86952.1 GL888237 EGI64216.1 CP012523 ALC39119.1 KQ976565 KYM80511.1 ADTU01002869 KQ981215 KYN44545.1
OWR46930.1 KZ149944 PZC76847.1 KQ459299 KPJ01915.1 RSAL01000173 RVE45081.1 JTDY01006012 KOB66374.1 NEVH01027059 PNF14009.1 PNF14012.1 PYGN01000836 PSN40194.1 KR733678 AKS04550.1 PNF14010.1 GBXI01000537 JAD13755.1 GBXI01014819 JAC99472.1 KK852868 KDR14696.1 GBXI01016770 JAC97521.1 GAMC01008935 JAB97620.1 GAKP01010198 JAC48754.1 GDHF01015917 JAI36397.1 GDHF01031692 JAI20622.1 GDHF01023613 JAI28701.1 GDHF01018633 JAI33681.1 GGFJ01002449 MBW51590.1 GDRN01030756 JAI67539.1 CH379064 KRT07119.1 GFDL01001285 JAV33760.1 GGFK01007050 MBW40371.1 GGFK01007059 MBW40380.1 CH954180 KQS30093.1 GGFM01003630 MBW24381.1 GGFJ01002024 MBW51165.1 GGFM01003710 MBW24461.1 GGFJ01002025 MBW51166.1 CH940655 KRF82554.1 CH933812 KRG07147.1 CH478141 EAT33821.1 ADMH02000350 ETN66868.1 GAMC01008933 JAB97622.1 OUUW01000031 SPP89791.1 SPP89790.1 GBXI01010759 JAD03533.1 CM000162 KRK06576.1 GDHF01005874 JAI46440.1 GAKP01010197 JAC48755.1 CCAG010022616 AE014298 ACZ95332.1 JRES01001467 KNC22653.1 AF118379 AY061148 AHN59914.1 LSMT01000338 PFX19929.1 CH964239 EDW82486.1 LJIG01016094 KRT81633.1 KQ971338 EFA02278.2 APCN01004264 AAAB01008960 EAA11766.4 GFDL01001281 JAV33764.1 OUUW01000002 SPP77053.1 QCYY01002451 ROT70212.1 GEZM01001085 GEZM01001084 JAV98171.1 ATLV01024126 KE525349 KFB50780.1 EAL32274.1 CH479193 EDW26897.1 GECZ01014644 JAS55125.1 AXCM01006421 UFQT01000888 SSX27846.1 AJFE02055016 AJFE02055017 AJFE02055018 AJFE02055019 JXUM01034315 JXUM01034316 JXUM01034317 KQ561019 KXJ80013.1 EAT33820.1 GECU01020320 JAS87386.1 EDV46710.2 JH167069 EHB00973.1 KN120575 KFO38223.1 EDX02127.1 KK107109 EZA59060.1 NNAY01002072 OXU22162.1 KQ977545 KYN02030.1 CVRI01000002 CRK86952.1 GL888237 EGI64216.1 CP012523 ALC39119.1 KQ976565 KYM80511.1 ADTU01002869 KQ981215 KYN44545.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000283053
UP000037510
UP000235965
+ More
UP000245037 UP000027135 UP000192221 UP000078200 UP000092445 UP000095301 UP000091820 UP000001819 UP000095300 UP000008711 UP000008792 UP000009192 UP000008820 UP000000673 UP000268350 UP000002282 UP000092444 UP000000803 UP000037069 UP000225706 UP000007798 UP000007266 UP000075840 UP000007062 UP000283509 UP000030765 UP000008744 UP000075885 UP000075883 UP000240080 UP000069940 UP000249989 UP000006813 UP000028990 UP000076408 UP000053097 UP000215335 UP000078542 UP000002358 UP000183832 UP000007755 UP000092553 UP000078540 UP000005205 UP000078541
UP000245037 UP000027135 UP000192221 UP000078200 UP000092445 UP000095301 UP000091820 UP000001819 UP000095300 UP000008711 UP000008792 UP000009192 UP000008820 UP000000673 UP000268350 UP000002282 UP000092444 UP000000803 UP000037069 UP000225706 UP000007798 UP000007266 UP000075840 UP000007062 UP000283509 UP000030765 UP000008744 UP000075885 UP000075883 UP000240080 UP000069940 UP000249989 UP000006813 UP000028990 UP000076408 UP000053097 UP000215335 UP000078542 UP000002358 UP000183832 UP000007755 UP000092553 UP000078540 UP000005205 UP000078541
Gene 3D
ProteinModelPortal
A0A2H1VKZ0
A0A2A4J4X6
A0A2A4J696
A0A212EZL8
A0A2W1BUG7
A0A194QA90
+ More
A0A3S2NUS6 A0A0L7KTL8 A0A2J7PCE5 A0A2J7PCF9 A0A2P8Y7R0 A0A187GBR9 A0A2J7PCE4 A0A0A1XS79 A0A0A1WM46 A0A067QYV0 A0A0A1WF42 W8B8N2 A0A034W254 A0A0K8VC34 A0A0K8U1N3 A0A0K8UQ43 A0A0K8V489 A0A1W4VVE2 A0A1A9VWY4 A0A1A9Z3W0 A0A1I8NA16 A0A2M4BEW0 A0A1A9W9K4 A0A0P4WIJ5 A0A0R3P5S0 A0A1I8PP14 A0A1Q3G1T4 A0A2M4AHX1 A0A2M4AHX5 A0A0Q5T4B1 A0A2M3Z798 A0A2M4BDP6 A0A2M3Z7H8 A0A2M4BDN9 A0A0Q9WDN2 A0A0Q9XF36 Q16HU3 W5JU97 W8BFW3 A0A3B0KWP9 A0A3B0KMD8 A0A0A1WYP6 A0A0R1EAT5 A0A1A9WFR2 A0A0K8W6D2 A0A034W3R9 A0A1B0G596 A0A1W4VHA8 E1JJQ3 A0A0L0BRN3 Q9XYP7-2 X2JG40 A0A1I8NA10 A0A1I8NA08 A0A2B4RSY1 B4NCC7 A0A0T6B308 D2A387 A0A3F2YSR6 Q7Q703 A0A1Q3G1T0 A0A3B0J9L1 A0A3R7M2R8 A0A1Y1NJV3 A0A084WKN6 Q29G36 B4GW52 A0A182PHG5 A0A1B6FY55 A0A182M1W1 A0A336MC89 A0A2R9BTL5 A0A182HDF5 Q16HU4 A0A1B6IKE5 B3NVP5 G5AVB2 A0A091E667 B4PZH1 A0A182Y143 A0A026WSN5 A0A232EUY2 A0A151IHY4 K7IYU5 A0A1J1HG27 F4WNM3 A0A0M3QTM0 A0A195B7L9 A0A158NUW2 A0A195FV78
A0A3S2NUS6 A0A0L7KTL8 A0A2J7PCE5 A0A2J7PCF9 A0A2P8Y7R0 A0A187GBR9 A0A2J7PCE4 A0A0A1XS79 A0A0A1WM46 A0A067QYV0 A0A0A1WF42 W8B8N2 A0A034W254 A0A0K8VC34 A0A0K8U1N3 A0A0K8UQ43 A0A0K8V489 A0A1W4VVE2 A0A1A9VWY4 A0A1A9Z3W0 A0A1I8NA16 A0A2M4BEW0 A0A1A9W9K4 A0A0P4WIJ5 A0A0R3P5S0 A0A1I8PP14 A0A1Q3G1T4 A0A2M4AHX1 A0A2M4AHX5 A0A0Q5T4B1 A0A2M3Z798 A0A2M4BDP6 A0A2M3Z7H8 A0A2M4BDN9 A0A0Q9WDN2 A0A0Q9XF36 Q16HU3 W5JU97 W8BFW3 A0A3B0KWP9 A0A3B0KMD8 A0A0A1WYP6 A0A0R1EAT5 A0A1A9WFR2 A0A0K8W6D2 A0A034W3R9 A0A1B0G596 A0A1W4VHA8 E1JJQ3 A0A0L0BRN3 Q9XYP7-2 X2JG40 A0A1I8NA10 A0A1I8NA08 A0A2B4RSY1 B4NCC7 A0A0T6B308 D2A387 A0A3F2YSR6 Q7Q703 A0A1Q3G1T0 A0A3B0J9L1 A0A3R7M2R8 A0A1Y1NJV3 A0A084WKN6 Q29G36 B4GW52 A0A182PHG5 A0A1B6FY55 A0A182M1W1 A0A336MC89 A0A2R9BTL5 A0A182HDF5 Q16HU4 A0A1B6IKE5 B3NVP5 G5AVB2 A0A091E667 B4PZH1 A0A182Y143 A0A026WSN5 A0A232EUY2 A0A151IHY4 K7IYU5 A0A1J1HG27 F4WNM3 A0A0M3QTM0 A0A195B7L9 A0A158NUW2 A0A195FV78
Ontologies
KEGG
GO
PANTHER
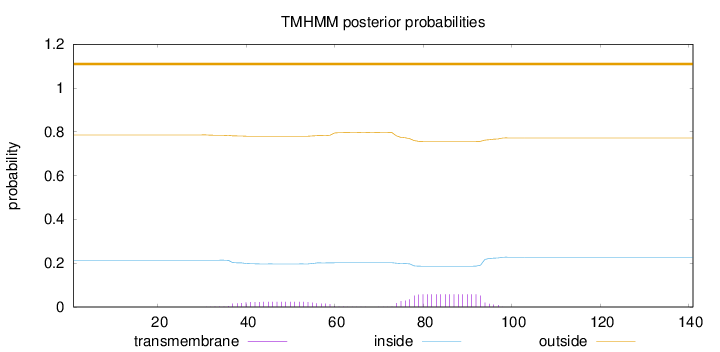
Topology
Subcellular location
Length:
141
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.57619
Exp number, first 60 AAs:
0.48837
Total prob of N-in:
0.21399
outside
1 - 141
Population Genetic Test Statistics
Pi
138.289276
Theta
158.534417
Tajima's D
-1.121619
CLR
236.969328
CSRT
0.11704414779261
Interpretation
Uncertain
