Pre Gene Modal
BGIBMGA001166
Annotation
transient_receptor_potential_cation_channel_protein_painless_[Bombyx_mori]
Full name
Transient receptor potential cation channel protein painless
Location in the cell
PlasmaMembrane Reliability : 2.476
Sequence
CDS
ATGTCACGTGATAATTACGAAATGAAGCCGAAGCAATTATCAAGAAACAGCTCTTTATTTGGAACCGATCCCCAAGAGCTGTTGAACAAGGCCCTTCAGAATAACGACTACGCGAAGTTCAAGAAACTCGTTACCGAAGCTAATGTTGATCTCGAACATGTTTATGAATATCCTGATTACAAAACATGCCTCGAAGTGGCTGTATCCGACAGAAACAAATTAGAATTCGTCAAATTACTATTACAATATCAAGTAGAAGTGAACAAAGTGAACGTTACACATTCTGCCGCACCCATACACTTTGCTGTCGAAAATGGCAACATAGACGCACTTGCGGCCCTTTTAGAAGACGACCGAATTGATGTTAACGTTAAAAGTAGGGGGAACACCGCGATATTGATGGCCGTCAAACAAATTGAAGAACTAGACGATGCCCGTGAACACGAATTAACAATATATGAAGATATGATTGAACTACTTCTAAAAGCTGGATGTAATGCAAATTCACCAGACCTTAAGGGTGTAACTCCTATTTATAGTGCAGCGAAACAAGGCCTAGAAAGAGTTATCACACTAATTATTGACTATGCAAAGCACGCTATTGACATAGACACGTACAAAGATAGAAGGGGTAAAACTGCTCGCGATTATTTAAAAGAAGCTTTCCCGTATCTAGAAGCAAAGTTTGATTCGACTACTCAAGACCCTGAAATTGTAGACTCTGATAAATTATTCTCATATTTAAGCAGACACGAAGAGGACAACTTTATAAGAGATTTTCTAAAACTTGCTAACAAAAACGAACATCGTAAAATATTGCCGGTCAACAATGGTATGAATACTATGTTACAACTAGCAACAGAGAAAGGTTTTGAAAAAGCAGTCAGTACTTTATTAAGATATGGGGCGGATCCTAACGCAACTTGTTCCAGTAATACGAGCAGACCGATTGCGTTGGCGTGTCAAAACGGATATTACAAAATTGTTAAAATGTTCATGGATAATGAGTCGACTTTGTTCGATCCAGTTAATTCGGAATCTCTAGTGCAAATTACTATAAAAGGAAGGCGCTCATCTATAAATATCCCTAATGTTAATTTTGATGAATGTTTGCGACTATTACTCAATCATCCTAAAATGGACATTAATGTTAATCATATGGATATGAAAAATAACACGGCCCTTCACTATGCTGCAAGAAACGGTGATAATAAAACTGTCCTGGAACTACTTAGAAACGGGGCTTGCATAGGATTGCGTAACTCGTTCGACGAACCACCACTCGCAGATATTAATGCTAAAACATTGGAAGCATTCTTGGATGAATGTGTCACAACGAACAACGAAAGACCAAGCAATGACGATTATGAAATACACATGAAATACAGTTTCCTAGTTTATCCAAATAATTCGCTAGAAAACGAACTTTGTAAAGTGCCATTAATCGACAATACTAACAATAACGTTAAAGAGTATGATACTATATTAGCACCCGAAACTGATGCTTTGTTATACATGACACGAAATGAAGAATTACGACCATTGCTAAAACACCCGGTGATAACAAGTTTTCTTTATTTAAAATGGCAAAGAATAAGCTGTCTTTTTTTTGCGAACATAACGTTTTATTCCTTTTTGTGGTTATGTTTAATTCTCTATATTATTTTGGGATACGGCGCTGAAAAGAAGCAACGAGACTCTTTTGAGGCGTTAAACGTTATCACACATGTCGGTGTTATTATTGGTGTGGTTTTGCTATTGATCAGGGAATTATTTCAACTTTTACTATCACCGACAAGATATTTACAGAGCATTGAAAATTGGATGGAAATCGGTCTTATTTTTGTGACGATTTGGATAGTGTTTTCTAAATCGGCTACAGAGTCGACGAAGCAGCAACTTTCAGCGGTTGCAATTTTGTTATCATCAGCCGAATTAGTTCTACTAATTGGACAGTTTCCGACACTGTCAACTAACATCGTCATGTTGAAAACGGTTTCGTGGAATTTCTTCAAATTTTTACTTTGGTATTGCATTCTCATAATAGCTTTTGCGTTAAGTTTCTACACACTATTCAGGCAAGAAAATGAAGAAGATCAGCGAGCACCAGACCCCAACACAGTCGGTAAAGAGGAGGAAGAAGAAGATTTCTTCGAAGATCCTGGTAGATCTTTATTTAAGACCATCGTCATGTTAACTGGAGAATTCGATGCATCTTCCATAAAATTCAGTACATATCCCTTAACGAGTCACATTATATTCATCGTGTTCGTTTTCATGATACCCATTGTGCTTTTTAATTTACTCAACGGTCTTGCGGTTAGTGACACACAAGAAATTAGGGCAGATGCCGAATTAGTAGGGCATATTTCTCGAATTAAACTTATTTCTTATATCGAGAGCGTGCTTATCGGTAGCGCGAAAACCCATTCACGACCATCAAAGTGTTGGTCGTTGCTGCCGTTCAACATTCAAAACTTAAATCTCATCAAACCTAAAACCTTTTTTACGAAATCCTTTGCAAAGAGGATTTGTTTGTTTCCCCATTTCCTGCCGAAATATAAAATACTTGTTAAACCAAATCAAAACAATATAATTGAGATACCGCACGCCGATGACAAAGACCTTGAAAGGAGCGGGGGGTGTTGTTTCGAGCGCTGTCAAAACTATAGGCTGGATCGTAAAATTGTCAAAAACGCAAAGCTAGTGATTAGTAATAAAACAAAGATAACCGAATTCGATGATAAGCTTTCGATGTACGAAAATAAAATTGAGAGTCTCGAGGCAACTCTGAAGAAAGTATTATTAGCCATTGAATCGCAACGCAATTAA
Protein
MSRDNYEMKPKQLSRNSSLFGTDPQELLNKALQNNDYAKFKKLVTEANVDLEHVYEYPDYKTCLEVAVSDRNKLEFVKLLLQYQVEVNKVNVTHSAAPIHFAVENGNIDALAALLEDDRIDVNVKSRGNTAILMAVKQIEELDDAREHELTIYEDMIELLLKAGCNANSPDLKGVTPIYSAAKQGLERVITLIIDYAKHAIDIDTYKDRRGKTARDYLKEAFPYLEAKFDSTTQDPEIVDSDKLFSYLSRHEEDNFIRDFLKLANKNEHRKILPVNNGMNTMLQLATEKGFEKAVSTLLRYGADPNATCSSNTSRPIALACQNGYYKIVKMFMDNESTLFDPVNSESLVQITIKGRRSSINIPNVNFDECLRLLLNHPKMDINVNHMDMKNNTALHYAARNGDNKTVLELLRNGACIGLRNSFDEPPLADINAKTLEAFLDECVTTNNERPSNDDYEIHMKYSFLVYPNNSLENELCKVPLIDNTNNNVKEYDTILAPETDALLYMTRNEELRPLLKHPVITSFLYLKWQRISCLFFANITFYSFLWLCLILYIILGYGAEKKQRDSFEALNVITHVGVIIGVVLLLIRELFQLLLSPTRYLQSIENWMEIGLIFVTIWIVFSKSATESTKQQLSAVAILLSSAELVLLIGQFPTLSTNIVMLKTVSWNFFKFLLWYCILIIAFALSFYTLFRQENEEDQRAPDPNTVGKEEEEEDFFEDPGRSLFKTIVMLTGEFDASSIKFSTYPLTSHIIFIVFVFMIPIVLFNLLNGLAVSDTQEIRADAELVGHISRIKLISYIESVLIGSAKTHSRPSKCWSLLPFNIQNLNLIKPKTFFTKSFAKRICLFPHFLPKYKILVKPNQNNIIEIPHADDKDLERSGGCCFERCQNYRLDRKIVKNAKLVISNKTKITEFDDKLSMYENKIESLEATLKKVLLAIESQRN
Summary
Description
Receptor-activated non-selective cation channel involved in detection of pain sensation due to high temperature. Involved in heat nociception by being activated by noxious temperature of 38 degrees Celsius.
Similarity
Belongs to the transient receptor (TC 1.A.4) family.
Keywords
ANK repeat
Complete proteome
Ion channel
Ion transport
Membrane
Reference proteome
Repeat
Sensory transduction
Transmembrane
Transmembrane helix
Transport
Feature
chain Transient receptor potential cation channel protein painless
Uniprot
H9IV86
W8VLJ7
A0A0L7KV58
A0A194Q921
A0A0N1IB44
A0A2W1BPE4
+ More
A0A2A4J6X8 A0A212FGX5 A0A2H1V5D0 A0A1B6CGD6 A0A384TDJ9 A0A1B6KZ14 A0A1B6JWY2 A0A1B6F0M2 A0A2P8Z590 A0A2J7QDV1 A0A2J7QDV3 A0A067R3X9 A0A2H8TFV4 A0A1Y1M4Y6 A0A1Y1M7K7 A0A2S2PRU0 A0A2S2R3K0 J9K079 A0A1W4XWE3 T1H8V5 A0A0A9YSC7 A0A0K8SLJ5 A0A023F0T9 A0A0C9RIG0 E2BX95 A0A348G651 A0A067QX85 A0A2R7VY74 A0A2P8YT24 A0A3L8DGE4 A0A2J7PS48 D2A388 A0A0J7L592 A0A026WVC4 A0A195FXH2 A0A1B0D8H9 A0A195ED38 A0A151XBZ5 F4WNM4 A0A195B8E2 E2AFM2 A0A151IHW4 A0A154PSN7 A0A088A461 A0A310SML5 A0A0M9A5P8 A0A0L7R634 A0A232EUY7 U5EX02 A0A0K2TXL7 Q174M7 A0A1W7R6N9 B3NR12 A0A0M4EUM6 A0A1S4FES5 A0A0P5J7P1 W8AWL1 A0A067R6H8 A0A034WER2 A0A0K8VZT7 A0A0L0BPE1 B4PBX3 B4QCM3 A0A0P6IL64 Q9W0Y6 E9G8Q1 B4MJW8 A0A0K2U030 B4J8W6 B3MC48 A0A182J552 B4IHA7 B4MEC5 A0A3B0J123 B4GD22 B0WK88 B5DZ25 A0A0P4W7B1 A0A0P6GNN0 A0A336N1Y4 A0A336LPG2 A0A1W4UZ98 A0A0N8AAY9 A0A164QBG9 A0A0A1XPF7 A0A0N8BX55 A0A182WZG9 Q174M6 A0A1A9WP89 A0A0P5LSY8 F5HIU3 A0A182HZL5
A0A2A4J6X8 A0A212FGX5 A0A2H1V5D0 A0A1B6CGD6 A0A384TDJ9 A0A1B6KZ14 A0A1B6JWY2 A0A1B6F0M2 A0A2P8Z590 A0A2J7QDV1 A0A2J7QDV3 A0A067R3X9 A0A2H8TFV4 A0A1Y1M4Y6 A0A1Y1M7K7 A0A2S2PRU0 A0A2S2R3K0 J9K079 A0A1W4XWE3 T1H8V5 A0A0A9YSC7 A0A0K8SLJ5 A0A023F0T9 A0A0C9RIG0 E2BX95 A0A348G651 A0A067QX85 A0A2R7VY74 A0A2P8YT24 A0A3L8DGE4 A0A2J7PS48 D2A388 A0A0J7L592 A0A026WVC4 A0A195FXH2 A0A1B0D8H9 A0A195ED38 A0A151XBZ5 F4WNM4 A0A195B8E2 E2AFM2 A0A151IHW4 A0A154PSN7 A0A088A461 A0A310SML5 A0A0M9A5P8 A0A0L7R634 A0A232EUY7 U5EX02 A0A0K2TXL7 Q174M7 A0A1W7R6N9 B3NR12 A0A0M4EUM6 A0A1S4FES5 A0A0P5J7P1 W8AWL1 A0A067R6H8 A0A034WER2 A0A0K8VZT7 A0A0L0BPE1 B4PBX3 B4QCM3 A0A0P6IL64 Q9W0Y6 E9G8Q1 B4MJW8 A0A0K2U030 B4J8W6 B3MC48 A0A182J552 B4IHA7 B4MEC5 A0A3B0J123 B4GD22 B0WK88 B5DZ25 A0A0P4W7B1 A0A0P6GNN0 A0A336N1Y4 A0A336LPG2 A0A1W4UZ98 A0A0N8AAY9 A0A164QBG9 A0A0A1XPF7 A0A0N8BX55 A0A182WZG9 Q174M6 A0A1A9WP89 A0A0P5LSY8 F5HIU3 A0A182HZL5
Pubmed
19121390
24639527
26227816
26354079
28756777
22118469
+ More
29403074 24845553 28004739 25401762 26823975 25474469 20798317 30249741 18362917 19820115 24508170 21719571 28648823 17510324 17994087 24495485 25348373 26108605 17550304 22936249 12705873 10731132 12537572 21292972 15632085 25830018 12364791 14747013 17210077
29403074 24845553 28004739 25401762 26823975 25474469 20798317 30249741 18362917 19820115 24508170 21719571 28648823 17510324 17994087 24495485 25348373 26108605 17550304 22936249 12705873 10731132 12537572 21292972 15632085 25830018 12364791 14747013 17210077
EMBL
BABH01000690
AB264789
BAO53208.1
JTDY01005527
KOB66914.1
KQ459299
+ More
KPJ01914.1 KQ460202 KPJ17180.1 KZ149944 PZC76848.1 NWSH01002968 PCG67200.1 AGBW02008583 OWR52996.1 ODYU01000763 SOQ36037.1 GEDC01024817 JAS12481.1 KX249693 AOR81470.1 GEBQ01023332 JAT16645.1 GECU01004017 JAT03690.1 GECZ01026128 JAS43641.1 PYGN01000189 PSN51663.1 NEVH01015683 PNF26761.1 PNF26762.1 KK852775 KDR16796.1 GFXV01000797 MBW12602.1 GEZM01041195 JAV80581.1 GEZM01041196 JAV80580.1 GGMR01019429 MBY32048.1 GGMS01015137 MBY84340.1 ABLF02020407 ACPB03006508 KU356754 GBHO01008520 GDHC01018488 APB88050.1 JAG35084.1 JAQ00141.1 GBRD01012159 JAG53665.1 GBBI01004091 JAC14621.1 GBYB01013010 GBYB01013011 GBYB01013012 JAG82777.1 JAG82778.1 JAG82779.1 GL451230 EFN79678.1 FX985591 BBF97924.1 KK852854 KDR14974.1 KK854155 PTY12279.1 PYGN01000377 PSN47403.1 QOIP01000008 RLU19517.1 NEVH01021939 PNF19162.1 KQ971338 EFA02836.1 LBMM01000695 KMQ97718.1 KK107109 EZA59059.1 KQ981215 KYN44544.1 AJVK01027446 KQ979074 KYN22759.1 KQ982316 KYQ57818.1 GL888237 EGI64217.1 KQ976565 KYM80512.1 GL439118 EFN67773.1 KQ977545 KYN02029.1 KQ435066 KZC14334.1 KU532290 ANT80565.1 KQ762962 OAD55186.1 KQ435728 KOX77755.1 KQ414648 KOC66313.1 NNAY01002072 OXU22161.1 GANO01000313 JAB59558.1 HACA01013224 CDW30585.1 CH477408 EAT41530.1 GEHC01000819 JAV46826.1 CH954179 EDV57095.1 CP012524 ALC41437.1 GDIQ01202002 JAK49723.1 GAMC01016088 JAB90467.1 KDR14973.1 GAKP01004881 JAC54071.1 GDHF01008199 JAI44115.1 JRES01001578 KNC21853.1 CM000158 EDW92627.1 CM000362 CM002911 EDX08639.1 KMY96473.1 GDIQ01029808 JAN64929.1 AY268106 AE013599 BT011483 GL732535 EFX84229.1 CH963846 EDW72407.1 HACA01014079 CDW31440.1 CH916367 EDW02406.1 CH902619 EDV37235.1 CH480838 EDW49283.1 CH940662 EDW58890.2 OUUW01000001 SPP74495.1 CH479181 EDW31560.1 DS231968 EDS29662.1 CM000071 EDY68761.1 GDRN01085983 JAI61269.1 GDIQ01031001 JAN63736.1 UFQT01002005 SSX32378.1 UFQT01000040 SSX18573.1 GDIP01165225 JAJ58177.1 LRGB01002451 KZS07610.1 GBXI01001396 JAD12896.1 GDIQ01136219 JAL15507.1 EAT41531.1 GDIQ01165709 JAK86016.1 AAAB01008799 EGK96204.1 APCN01001985
KPJ01914.1 KQ460202 KPJ17180.1 KZ149944 PZC76848.1 NWSH01002968 PCG67200.1 AGBW02008583 OWR52996.1 ODYU01000763 SOQ36037.1 GEDC01024817 JAS12481.1 KX249693 AOR81470.1 GEBQ01023332 JAT16645.1 GECU01004017 JAT03690.1 GECZ01026128 JAS43641.1 PYGN01000189 PSN51663.1 NEVH01015683 PNF26761.1 PNF26762.1 KK852775 KDR16796.1 GFXV01000797 MBW12602.1 GEZM01041195 JAV80581.1 GEZM01041196 JAV80580.1 GGMR01019429 MBY32048.1 GGMS01015137 MBY84340.1 ABLF02020407 ACPB03006508 KU356754 GBHO01008520 GDHC01018488 APB88050.1 JAG35084.1 JAQ00141.1 GBRD01012159 JAG53665.1 GBBI01004091 JAC14621.1 GBYB01013010 GBYB01013011 GBYB01013012 JAG82777.1 JAG82778.1 JAG82779.1 GL451230 EFN79678.1 FX985591 BBF97924.1 KK852854 KDR14974.1 KK854155 PTY12279.1 PYGN01000377 PSN47403.1 QOIP01000008 RLU19517.1 NEVH01021939 PNF19162.1 KQ971338 EFA02836.1 LBMM01000695 KMQ97718.1 KK107109 EZA59059.1 KQ981215 KYN44544.1 AJVK01027446 KQ979074 KYN22759.1 KQ982316 KYQ57818.1 GL888237 EGI64217.1 KQ976565 KYM80512.1 GL439118 EFN67773.1 KQ977545 KYN02029.1 KQ435066 KZC14334.1 KU532290 ANT80565.1 KQ762962 OAD55186.1 KQ435728 KOX77755.1 KQ414648 KOC66313.1 NNAY01002072 OXU22161.1 GANO01000313 JAB59558.1 HACA01013224 CDW30585.1 CH477408 EAT41530.1 GEHC01000819 JAV46826.1 CH954179 EDV57095.1 CP012524 ALC41437.1 GDIQ01202002 JAK49723.1 GAMC01016088 JAB90467.1 KDR14973.1 GAKP01004881 JAC54071.1 GDHF01008199 JAI44115.1 JRES01001578 KNC21853.1 CM000158 EDW92627.1 CM000362 CM002911 EDX08639.1 KMY96473.1 GDIQ01029808 JAN64929.1 AY268106 AE013599 BT011483 GL732535 EFX84229.1 CH963846 EDW72407.1 HACA01014079 CDW31440.1 CH916367 EDW02406.1 CH902619 EDV37235.1 CH480838 EDW49283.1 CH940662 EDW58890.2 OUUW01000001 SPP74495.1 CH479181 EDW31560.1 DS231968 EDS29662.1 CM000071 EDY68761.1 GDRN01085983 JAI61269.1 GDIQ01031001 JAN63736.1 UFQT01002005 SSX32378.1 UFQT01000040 SSX18573.1 GDIP01165225 JAJ58177.1 LRGB01002451 KZS07610.1 GBXI01001396 JAD12896.1 GDIQ01136219 JAL15507.1 EAT41531.1 GDIQ01165709 JAK86016.1 AAAB01008799 EGK96204.1 APCN01001985
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000218220
UP000007151
+ More
UP000245037 UP000235965 UP000027135 UP000007819 UP000192223 UP000015103 UP000008237 UP000279307 UP000007266 UP000036403 UP000053097 UP000078541 UP000092462 UP000078492 UP000075809 UP000007755 UP000078540 UP000000311 UP000078542 UP000076502 UP000005203 UP000053105 UP000053825 UP000215335 UP000008820 UP000008711 UP000092553 UP000037069 UP000002282 UP000000304 UP000000803 UP000000305 UP000007798 UP000001070 UP000007801 UP000075880 UP000001292 UP000008792 UP000268350 UP000008744 UP000002320 UP000001819 UP000192221 UP000076858 UP000076407 UP000091820 UP000007062 UP000075840
UP000245037 UP000235965 UP000027135 UP000007819 UP000192223 UP000015103 UP000008237 UP000279307 UP000007266 UP000036403 UP000053097 UP000078541 UP000092462 UP000078492 UP000075809 UP000007755 UP000078540 UP000000311 UP000078542 UP000076502 UP000005203 UP000053105 UP000053825 UP000215335 UP000008820 UP000008711 UP000092553 UP000037069 UP000002282 UP000000304 UP000000803 UP000000305 UP000007798 UP000001070 UP000007801 UP000075880 UP000001292 UP000008792 UP000268350 UP000008744 UP000002320 UP000001819 UP000192221 UP000076858 UP000076407 UP000091820 UP000007062 UP000075840
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
H9IV86
W8VLJ7
A0A0L7KV58
A0A194Q921
A0A0N1IB44
A0A2W1BPE4
+ More
A0A2A4J6X8 A0A212FGX5 A0A2H1V5D0 A0A1B6CGD6 A0A384TDJ9 A0A1B6KZ14 A0A1B6JWY2 A0A1B6F0M2 A0A2P8Z590 A0A2J7QDV1 A0A2J7QDV3 A0A067R3X9 A0A2H8TFV4 A0A1Y1M4Y6 A0A1Y1M7K7 A0A2S2PRU0 A0A2S2R3K0 J9K079 A0A1W4XWE3 T1H8V5 A0A0A9YSC7 A0A0K8SLJ5 A0A023F0T9 A0A0C9RIG0 E2BX95 A0A348G651 A0A067QX85 A0A2R7VY74 A0A2P8YT24 A0A3L8DGE4 A0A2J7PS48 D2A388 A0A0J7L592 A0A026WVC4 A0A195FXH2 A0A1B0D8H9 A0A195ED38 A0A151XBZ5 F4WNM4 A0A195B8E2 E2AFM2 A0A151IHW4 A0A154PSN7 A0A088A461 A0A310SML5 A0A0M9A5P8 A0A0L7R634 A0A232EUY7 U5EX02 A0A0K2TXL7 Q174M7 A0A1W7R6N9 B3NR12 A0A0M4EUM6 A0A1S4FES5 A0A0P5J7P1 W8AWL1 A0A067R6H8 A0A034WER2 A0A0K8VZT7 A0A0L0BPE1 B4PBX3 B4QCM3 A0A0P6IL64 Q9W0Y6 E9G8Q1 B4MJW8 A0A0K2U030 B4J8W6 B3MC48 A0A182J552 B4IHA7 B4MEC5 A0A3B0J123 B4GD22 B0WK88 B5DZ25 A0A0P4W7B1 A0A0P6GNN0 A0A336N1Y4 A0A336LPG2 A0A1W4UZ98 A0A0N8AAY9 A0A164QBG9 A0A0A1XPF7 A0A0N8BX55 A0A182WZG9 Q174M6 A0A1A9WP89 A0A0P5LSY8 F5HIU3 A0A182HZL5
A0A2A4J6X8 A0A212FGX5 A0A2H1V5D0 A0A1B6CGD6 A0A384TDJ9 A0A1B6KZ14 A0A1B6JWY2 A0A1B6F0M2 A0A2P8Z590 A0A2J7QDV1 A0A2J7QDV3 A0A067R3X9 A0A2H8TFV4 A0A1Y1M4Y6 A0A1Y1M7K7 A0A2S2PRU0 A0A2S2R3K0 J9K079 A0A1W4XWE3 T1H8V5 A0A0A9YSC7 A0A0K8SLJ5 A0A023F0T9 A0A0C9RIG0 E2BX95 A0A348G651 A0A067QX85 A0A2R7VY74 A0A2P8YT24 A0A3L8DGE4 A0A2J7PS48 D2A388 A0A0J7L592 A0A026WVC4 A0A195FXH2 A0A1B0D8H9 A0A195ED38 A0A151XBZ5 F4WNM4 A0A195B8E2 E2AFM2 A0A151IHW4 A0A154PSN7 A0A088A461 A0A310SML5 A0A0M9A5P8 A0A0L7R634 A0A232EUY7 U5EX02 A0A0K2TXL7 Q174M7 A0A1W7R6N9 B3NR12 A0A0M4EUM6 A0A1S4FES5 A0A0P5J7P1 W8AWL1 A0A067R6H8 A0A034WER2 A0A0K8VZT7 A0A0L0BPE1 B4PBX3 B4QCM3 A0A0P6IL64 Q9W0Y6 E9G8Q1 B4MJW8 A0A0K2U030 B4J8W6 B3MC48 A0A182J552 B4IHA7 B4MEC5 A0A3B0J123 B4GD22 B0WK88 B5DZ25 A0A0P4W7B1 A0A0P6GNN0 A0A336N1Y4 A0A336LPG2 A0A1W4UZ98 A0A0N8AAY9 A0A164QBG9 A0A0A1XPF7 A0A0N8BX55 A0A182WZG9 Q174M6 A0A1A9WP89 A0A0P5LSY8 F5HIU3 A0A182HZL5
PDB
6MOL
E-value=1.45762e-13,
Score=189
Ontologies
GO
GO:0005216
GO:0016021
GO:0006811
GO:0050960
GO:0019233
GO:0042048
GO:0070588
GO:0005227
GO:0048060
GO:0045924
GO:0008016
GO:0007620
GO:0007638
GO:0050974
GO:0034704
GO:0008049
GO:0050966
GO:0050965
GO:0007631
GO:0009408
GO:0009612
GO:0035179
GO:0048266
GO:0005262
GO:0005886
GO:0006816
GO:0005515
GO:0016020
GO:0043015
GO:0007169
GO:0006412
GO:0016876
GO:0043039
GO:0055085
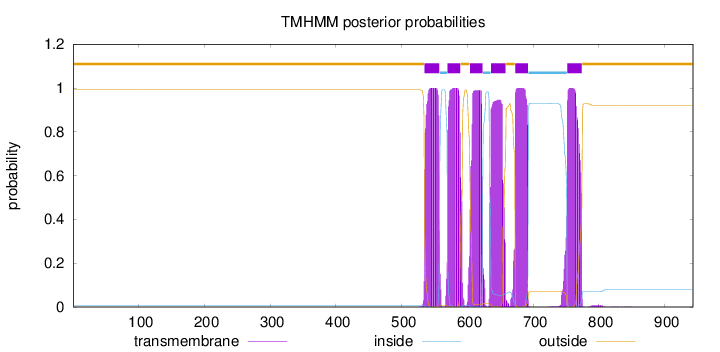
Topology
Subcellular location
Membrane
Length:
943
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
126.78375
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00729
outside
1 - 534
TMhelix
535 - 557
inside
558 - 569
TMhelix
570 - 589
outside
590 - 603
TMhelix
604 - 623
inside
624 - 635
TMhelix
636 - 658
outside
659 - 672
TMhelix
673 - 692
inside
693 - 751
TMhelix
752 - 774
outside
775 - 943
Population Genetic Test Statistics
Pi
160.017589
Theta
163.085172
Tajima's D
-0.463291
CLR
1.042381
CSRT
0.247987600619969
Interpretation
Uncertain
