Gene
KWMTBOMO07931
Pre Gene Modal
BGIBMGA001167
Annotation
PREDICTED:_transcription_factor_ETV6_[Amyelois_transitella]
Transcription factor
Location in the cell
Nuclear Reliability : 3.961
Sequence
CDS
ATGAAAGTTGTAAGCTTACAACTGCCGTCTGGGCCCAGTATGGAGCGGCTACCGCTACCATTCAGTCCAACAGAACTATTATGGCGTTACCCACTACCCTGGGCGCCACCGCCTCCTTCACCGCTTGGTGACACCAAGGCGCAACTGCCAGCCGGTCTTCCGCCAGAACCAAGACTATGGACCAGAGAGGATGTTTCTGTATTTCTGAAATGGTGTGAAAGAGAATTCGATCTACCTAATTTTGATATGGACCTCTTTCAAATGAACGGTAAAGCATTATGCTTGCTTACTAAAACAGATTTGGGTGAAAGGTGTCCTGGAGCAGGCGATGTCTTACATAATGTTCTTCAAATGCTTGTACGCGACGCAGCGCTGTTAGGTAGAGTTCCTTCCTCACCTGTTACTCCCACCGCTCGTGCGGCACCGTATCCGCCCTCTCCCCACTCTCATCCACCGACACCAACTTGGACTGTTGATGGTTTCCACCATTTCCACAGCGCTGCAGCAGCAGCAGCAGCTCAACCTAATTCAGTGACTCTCAGTCCTGCTCCATCAGTTGATAGTTCTGGAAGTCCTCAAAGAGGAGACACTGTAACTTATGCGCCAGCGTATGCACCGTCCGTGCCCCCCACTACTCAAGCTGCCAGTTCCGGAAGTAATCATTCAGATTCCGATGAAGAGGGTCAATACGCTCCACCTCCACGTTCTCCTAAAGAAGCTCCAATAACGAGTCCAGCCCCGCAAAATCATCCAACACAACAACATCCTCATTACCGCGCTCAACATAGGGAATTTTTTCCAAATGATATGCCAGAGTCCAATACAAATGGAAGACTATTGTGGGACTTCTTACAACAACTTCTGAATGACCCAACGCAAAGATACACAAACTATATTGCATGGAAGAACAGAGACACTGGCGTGTTTAAAATTGTTGATCCTGCTGGACTAGCCAAATTGTGGGGAATTCAAAAAAATCACCTATCCATGAACTACGACAAAATGTCCCGCGCCCTGAGATATTATTACCGTGTTAATATTTTGCGCAAAGTCCAGGGCGAAAGACACTGTTACCAGTTCTTAAGAAACCCAACAGAATTGAAAAATATCAAAAACATATCTCTACTGCGACAACAAATGAGTCCCACTCGCGTTGTACCACAAACTTTGGTAAAAACCGAAATGAAAGAAGAAAGATGTGACGAAGAAACAGTCGATGAAGAAATGCCTACAGATTTGAGTATGTCTGCATCAGAGCCTTGGCGGAAACGCGCACGCTCTGACACAGCGACCGCACCGCCGTCGTCAACGCAACATGACAAACATCGGATCAGTACGCTTATCGGAGATAACATGATAATGAAACGCGAAGTCGACTACAGTGCTGAGCATTATGCCTTAAATTTAAAAAGTGAAAAATGTGAACAGTAA
Protein
MKVVSLQLPSGPSMERLPLPFSPTELLWRYPLPWAPPPPSPLGDTKAQLPAGLPPEPRLWTREDVSVFLKWCEREFDLPNFDMDLFQMNGKALCLLTKTDLGERCPGAGDVLHNVLQMLVRDAALLGRVPSSPVTPTARAAPYPPSPHSHPPTPTWTVDGFHHFHSAAAAAAAQPNSVTLSPAPSVDSSGSPQRGDTVTYAPAYAPSVPPTTQAASSGSNHSDSDEEGQYAPPPRSPKEAPITSPAPQNHPTQQHPHYRAQHREFFPNDMPESNTNGRLLWDFLQQLLNDPTQRYTNYIAWKNRDTGVFKIVDPAGLAKLWGIQKNHLSMNYDKMSRALRYYYRVNILRKVQGERHCYQFLRNPTELKNIKNISLLRQQMSPTRVVPQTLVKTEMKEERCDEETVDEEMPTDLSMSASEPWRKRARSDTATAPPSSTQHDKHRISTLIGDNMIMKREVDYSAEHYALNLKSEKCEQ
Summary
Similarity
Belongs to the ETS family.
Uniprot
A0A060MZU1
H9IV87
A0A3S2LV95
A0A2A4JWA2
A0A2H1VVT5
A0A194Q8R3
+ More
A0A0N1PHP8 A0A2W1BT73 A0A212ETP5 S4P5B6 A0A0L7LNR2 D2A2D2 A0A1Y1L4A1 A0A067QWI9 A0A0M8ZRG0 A0A154PDW1 A0A2J7PCL4 A0A1W4WP27 A0A1W4WYT5 A0A0L7R7I4 A0A088A2Q8 A0A026W901 U4UFY4 N6TW97 A0A2P8Y7F9 A0A0C9QJM4 A0A1L8DI34 A0A1L8DI23 A0A1L8DI20 A0A195AYJ7 A0A151WXG8 A0A195CGK1 F4WAN5 A0A0J7L0D9 E9IM04 A0A195F9A0 E2A4E3 E2C7L7 A0A3Q0JEU3 A0A1B6KJT3 E0VDJ0 A0A0A9X314 A0A023F2Y3 A0A0T6ATS8 A0A069DZ83 A0A1B6F1I1 A0A195DYE5 A0A0A1X212 A0A0K8V5K1 A0A0K8TXD2 T1I5Z9 A0A1B6J2W9 A0A1B0FPR5 A0A2H8TX82 A0A2S2PS18 A0A1B6EXL1 A0A2S2QNZ1 X1WJF4 A0A1B6EJ34 A0A226E968 A0A0P4W945 A0A0K8TW77 A0A1Z5KUT9 A0A293LMT3
A0A0N1PHP8 A0A2W1BT73 A0A212ETP5 S4P5B6 A0A0L7LNR2 D2A2D2 A0A1Y1L4A1 A0A067QWI9 A0A0M8ZRG0 A0A154PDW1 A0A2J7PCL4 A0A1W4WP27 A0A1W4WYT5 A0A0L7R7I4 A0A088A2Q8 A0A026W901 U4UFY4 N6TW97 A0A2P8Y7F9 A0A0C9QJM4 A0A1L8DI34 A0A1L8DI23 A0A1L8DI20 A0A195AYJ7 A0A151WXG8 A0A195CGK1 F4WAN5 A0A0J7L0D9 E9IM04 A0A195F9A0 E2A4E3 E2C7L7 A0A3Q0JEU3 A0A1B6KJT3 E0VDJ0 A0A0A9X314 A0A023F2Y3 A0A0T6ATS8 A0A069DZ83 A0A1B6F1I1 A0A195DYE5 A0A0A1X212 A0A0K8V5K1 A0A0K8TXD2 T1I5Z9 A0A1B6J2W9 A0A1B0FPR5 A0A2H8TX82 A0A2S2PS18 A0A1B6EXL1 A0A2S2QNZ1 X1WJF4 A0A1B6EJ34 A0A226E968 A0A0P4W945 A0A0K8TW77 A0A1Z5KUT9 A0A293LMT3
Pubmed
EMBL
AB510032
BAN37076.1
BABH01000697
RSAL01000173
RVE45083.1
NWSH01000456
+ More
PCG76281.1 ODYU01004731 SOQ44918.1 KQ459299 KPJ01918.1 KQ460202 KPJ17183.1 KZ149944 PZC76844.1 AGBW02012542 OWR44863.1 GAIX01007266 JAA85294.1 JTDY01000507 KOB76856.1 KQ971338 EFA02175.2 GEZM01071296 JAV65897.1 KK852868 KDR14699.1 KQ435951 KOX68123.1 KQ434869 KZC09370.1 NEVH01027059 PNF14073.1 KQ414642 KOC66726.1 KK107386 QOIP01000002 EZA51509.1 RLU25639.1 KB632337 ERL92864.1 APGK01014040 APGK01014041 KB739180 ENN82348.1 PYGN01000836 PSN40198.1 GBYB01003724 JAG73491.1 GFDF01007972 JAV06112.1 GFDF01007966 JAV06118.1 GFDF01007965 JAV06119.1 KQ976703 KYM77115.1 KQ982668 KYQ52506.1 KQ977791 KYM99842.1 GL888050 EGI68793.1 LBMM01001475 KMQ96252.1 GL764129 EFZ18294.1 KQ981727 KYN36787.1 GL436635 EFN71690.1 GL453383 EFN76093.1 GEBQ01028268 GEBQ01014498 JAT11709.1 JAT25479.1 DS235078 EEB11446.1 GBHO01029211 GBHO01029210 GBRD01016051 GBRD01005154 GDHC01003340 JAG14393.1 JAG14394.1 JAG49775.1 JAQ15289.1 GBBI01003139 JAC15573.1 LJIG01022871 KRT78299.1 GBGD01001090 JAC87799.1 GECZ01028615 GECZ01027771 GECZ01025613 GECZ01023654 GECZ01011833 GECZ01008740 GECZ01006358 GECZ01005359 JAS41154.1 JAS41998.1 JAS44156.1 JAS46115.1 JAS57936.1 JAS61029.1 JAS63411.1 JAS64410.1 KQ980066 KYN17903.1 GBXI01009130 JAD05162.1 GDHF01018454 JAI33860.1 GDHF01033373 JAI18941.1 ACPB03013074 GECU01014161 JAS93545.1 CCAG010019933 GFXV01005973 MBW17778.1 GGMR01019650 MBY32269.1 GECZ01027218 JAS42551.1 GGMS01010283 MBY79486.1 ABLF02012080 GECZ01031867 JAS37902.1 LNIX01000006 OXA53186.1 GDRN01079274 JAI62430.1 GDHF01033766 JAI18548.1 GFJQ02008107 JAV98862.1 GFWV01010068 MAA34797.1
PCG76281.1 ODYU01004731 SOQ44918.1 KQ459299 KPJ01918.1 KQ460202 KPJ17183.1 KZ149944 PZC76844.1 AGBW02012542 OWR44863.1 GAIX01007266 JAA85294.1 JTDY01000507 KOB76856.1 KQ971338 EFA02175.2 GEZM01071296 JAV65897.1 KK852868 KDR14699.1 KQ435951 KOX68123.1 KQ434869 KZC09370.1 NEVH01027059 PNF14073.1 KQ414642 KOC66726.1 KK107386 QOIP01000002 EZA51509.1 RLU25639.1 KB632337 ERL92864.1 APGK01014040 APGK01014041 KB739180 ENN82348.1 PYGN01000836 PSN40198.1 GBYB01003724 JAG73491.1 GFDF01007972 JAV06112.1 GFDF01007966 JAV06118.1 GFDF01007965 JAV06119.1 KQ976703 KYM77115.1 KQ982668 KYQ52506.1 KQ977791 KYM99842.1 GL888050 EGI68793.1 LBMM01001475 KMQ96252.1 GL764129 EFZ18294.1 KQ981727 KYN36787.1 GL436635 EFN71690.1 GL453383 EFN76093.1 GEBQ01028268 GEBQ01014498 JAT11709.1 JAT25479.1 DS235078 EEB11446.1 GBHO01029211 GBHO01029210 GBRD01016051 GBRD01005154 GDHC01003340 JAG14393.1 JAG14394.1 JAG49775.1 JAQ15289.1 GBBI01003139 JAC15573.1 LJIG01022871 KRT78299.1 GBGD01001090 JAC87799.1 GECZ01028615 GECZ01027771 GECZ01025613 GECZ01023654 GECZ01011833 GECZ01008740 GECZ01006358 GECZ01005359 JAS41154.1 JAS41998.1 JAS44156.1 JAS46115.1 JAS57936.1 JAS61029.1 JAS63411.1 JAS64410.1 KQ980066 KYN17903.1 GBXI01009130 JAD05162.1 GDHF01018454 JAI33860.1 GDHF01033373 JAI18941.1 ACPB03013074 GECU01014161 JAS93545.1 CCAG010019933 GFXV01005973 MBW17778.1 GGMR01019650 MBY32269.1 GECZ01027218 JAS42551.1 GGMS01010283 MBY79486.1 ABLF02012080 GECZ01031867 JAS37902.1 LNIX01000006 OXA53186.1 GDRN01079274 JAI62430.1 GDHF01033766 JAI18548.1 GFJQ02008107 JAV98862.1 GFWV01010068 MAA34797.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000007266 UP000027135 UP000053105 UP000076502 UP000235965 UP000192223 UP000053825 UP000005203 UP000053097 UP000279307 UP000030742 UP000019118 UP000245037 UP000078540 UP000075809 UP000078542 UP000007755 UP000036403 UP000078541 UP000000311 UP000008237 UP000079169 UP000009046 UP000078492 UP000015103 UP000092444 UP000007819 UP000198287
UP000037510 UP000007266 UP000027135 UP000053105 UP000076502 UP000235965 UP000192223 UP000053825 UP000005203 UP000053097 UP000279307 UP000030742 UP000019118 UP000245037 UP000078540 UP000075809 UP000078542 UP000007755 UP000036403 UP000078541 UP000000311 UP000008237 UP000079169 UP000009046 UP000078492 UP000015103 UP000092444 UP000007819 UP000198287
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A060MZU1
H9IV87
A0A3S2LV95
A0A2A4JWA2
A0A2H1VVT5
A0A194Q8R3
+ More
A0A0N1PHP8 A0A2W1BT73 A0A212ETP5 S4P5B6 A0A0L7LNR2 D2A2D2 A0A1Y1L4A1 A0A067QWI9 A0A0M8ZRG0 A0A154PDW1 A0A2J7PCL4 A0A1W4WP27 A0A1W4WYT5 A0A0L7R7I4 A0A088A2Q8 A0A026W901 U4UFY4 N6TW97 A0A2P8Y7F9 A0A0C9QJM4 A0A1L8DI34 A0A1L8DI23 A0A1L8DI20 A0A195AYJ7 A0A151WXG8 A0A195CGK1 F4WAN5 A0A0J7L0D9 E9IM04 A0A195F9A0 E2A4E3 E2C7L7 A0A3Q0JEU3 A0A1B6KJT3 E0VDJ0 A0A0A9X314 A0A023F2Y3 A0A0T6ATS8 A0A069DZ83 A0A1B6F1I1 A0A195DYE5 A0A0A1X212 A0A0K8V5K1 A0A0K8TXD2 T1I5Z9 A0A1B6J2W9 A0A1B0FPR5 A0A2H8TX82 A0A2S2PS18 A0A1B6EXL1 A0A2S2QNZ1 X1WJF4 A0A1B6EJ34 A0A226E968 A0A0P4W945 A0A0K8TW77 A0A1Z5KUT9 A0A293LMT3
A0A0N1PHP8 A0A2W1BT73 A0A212ETP5 S4P5B6 A0A0L7LNR2 D2A2D2 A0A1Y1L4A1 A0A067QWI9 A0A0M8ZRG0 A0A154PDW1 A0A2J7PCL4 A0A1W4WP27 A0A1W4WYT5 A0A0L7R7I4 A0A088A2Q8 A0A026W901 U4UFY4 N6TW97 A0A2P8Y7F9 A0A0C9QJM4 A0A1L8DI34 A0A1L8DI23 A0A1L8DI20 A0A195AYJ7 A0A151WXG8 A0A195CGK1 F4WAN5 A0A0J7L0D9 E9IM04 A0A195F9A0 E2A4E3 E2C7L7 A0A3Q0JEU3 A0A1B6KJT3 E0VDJ0 A0A0A9X314 A0A023F2Y3 A0A0T6ATS8 A0A069DZ83 A0A1B6F1I1 A0A195DYE5 A0A0A1X212 A0A0K8V5K1 A0A0K8TXD2 T1I5Z9 A0A1B6J2W9 A0A1B0FPR5 A0A2H8TX82 A0A2S2PS18 A0A1B6EXL1 A0A2S2QNZ1 X1WJF4 A0A1B6EJ34 A0A226E968 A0A0P4W945 A0A0K8TW77 A0A1Z5KUT9 A0A293LMT3
PDB
1SV4
E-value=1.80614e-28,
Score=314
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Nucleus
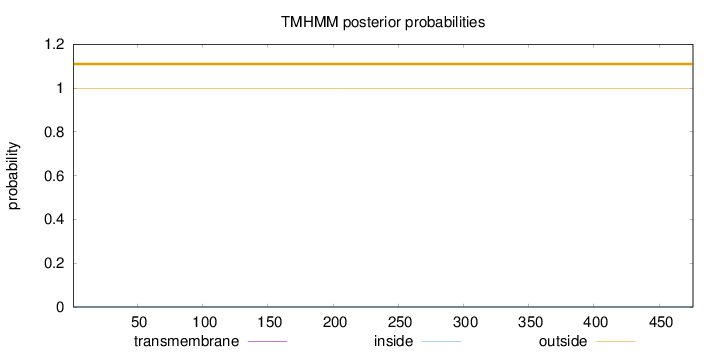
Length:
476
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00011
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00103
outside
1 - 476
Population Genetic Test Statistics
Pi
158.893224
Theta
184.368888
Tajima's D
-0.968712
CLR
0.694872
CSRT
0.143442827858607
Interpretation
Uncertain
