Gene
KWMTBOMO07920
Pre Gene Modal
BGIBMGA014468
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC106709759_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.37
Sequence
CDS
ATGGCACCCGTAATCCCTGAAATAACATTACCCGCGTCTGCAGAGGACATAAACAGTAGGGACACATACCTCGAAACCGTCCGTAAGCTCATATCCTGCATCACAACAGCTGTGACTGAGGGAAAAAGTGTTACCGCAGCTAACAAGCGCCTTATCTGCTCCGCGGCGGAGGAAATTCGTAGGGCGACGAGAGTGTTGGAAACAACACTCGCATCGACACTGACACCACCAACCCCCTCTCCACCTGCCACCAACGAAGGTTTGAAGGAGGAGATTATCGCTTGCGTCAGAGAGGAGTTCAAGCGGCATAGACAACTACCGCCGCTGACCCTTCAATCACAAGTTCGCCCATCGTATGCCCAAGCCGCAGCACCCAACCCCCGGAAACCAGCCCTTACGACCAAACCAACACCCAAAACATATCCAGCTACAAAACCTGCAATTATTGTCTCAGCAAAAACCGAAGTAAAGACGAAACAGGATATTATCGAGGCCTGGAGGCGCAACATCAGCTTCAAAAATTCCGGCTTTGCCCCTAGCAGAGTACAAGCTGTATCAAATAACAAACTAAGAATCGAGTTCGATAACATAGAACAAAGGAACGATACCCTCACTCGTATCACTAACTCCCAGACAATTCACGCCGAACCCGCTCGCAGGCTGCTGCCTATGCTCAATCTCAAGGGAGTCTCCAAAGACGTTCCCGCTGAAGATCTAGTGAGCATAATCAGCAGACAGAACCCGGAACTCACTAACGCAATCAACCAAAAGGAGGACTTGCGGTTTCGTTTCCAGCGCTCCAACAGAAACCCCAACTTATATAACGCCATCCTAATTGCACAGCCAACTGTCTGGCGTAAGTGTATAAGCCTGGGCAGAATCAGTGTGGACCATCAACGTATTCACGTAGAGGAGTTCACACCGTTCCTGCAGTGTCACAAGTGCCTTAGCTTCGGCCACACTAAAAACAAGTGTTCGTCCACCGAACAATACTGTGCACACTGCAGCGCAAATAACGAGGTCAAAAACATCCCCGGAATTAACATTTACCAATTCTCTGACGGCGCGCGTGTCAAGGCCTGCATACTAGCTAAGACAGGTGTCGGCTCAGTCCTCGGCCTATCCCAATACTCTTCGGCAAACCTGTCGGTCATACAGCTAACGGCAGGAAACAAGAAGACGTACATAGCCTCAACGTACATCGAACCCGATGTCGATGCCAACGGCACCATGGAGCGTCTGGAAGTTTTCCTTAAAGACATAGGAGGGGCGAGAGTAATTGTGGGAGGTGACTTCAATGGATGGCACCCATCGTGGGGGAGCGTGAGGGTGAACCCCAGAGGCTCATCACTAGTCGAACTAGCGCACAGCAACGATTTATACATTTGTAACTCAGGAGACACTCCAACCTTCGAAACCATCACTCATGGCCGCAATAGATCATCCATCATCGACCTTACTCTTGCATCCAGCGACCTCTTCAACAGCATCTCTGAATGGCACGTAAATCTTAACACATGCCCCACCTCCCAACACAACGCCATTGAATTCTCACTTAACACCAACGAAAACACAAATAAACACGCAGAAAACACTTCCACATTCCTATACAAGTCGGAACAAGCCAATTGGACAATATTTAGAAACACCCTCCACACTCTTATGGCCAACACAGACATCCTTGATAGGAGCGTTGAAAGCTTAAGCCCAGAGGGACTTGAGTCGCTCCTTCGGGAAGTCACGGACATCCTACACCGGGCCTGTAGGGCATCGATGCGAGTAAAAGCAGGCTCTGGGAATAGACACAAGCCGCCTTGGTGGAACGATCGTCTTCAGGACCTTAAAAACGAGGTAATCAACGTACATCGCAGATTACGTGCGGCCAAGAGACAGAACCTACCATTGGATGACCTCATAGAGCAACAGCAGACGAAAAAAGCCCTATACGCCGATGCCCTACGTGCAGAGTCGACGAGAAGCTTCCGCAGCTTCTGTGAATTACAAACGAAGGAAAATGTATGGTCACTCACCAACAGACTTTTAAAGGAATCCACCCCTAAACGGCCACCAGCTACTCTTAAGACTGGAAATACATTCACCACCTGTGCAAAAGAGACGGCGGAGGCTCTCTTAGAGCACTTTTACCCTGCTGACACTCCAGACACCATAGCACGACATCACCAACTTCGCGCATGCTACGACGCTCCCGTAGACACTGAAAATGACCTACCATTCACCGAGGAGGAGGTCATGGAGTGTCTGCAATACATGAACCCCAAAAAGGCCCCAGGGCTCGATCACATAACAGCTGACATATGCCAACAATTCGCATCCGCTTACCCCCGCTTAATCACCAACATTATGAACCGCTGCCTCACCCTTCAACACTTCCCCGATCAGTGGAAAACTGCGTATGTAAAAATCATCCCGAAGCCCGGTAAAAGCGACCACACATCTCTCACCTCATTCCGACCCATTGGACTCATCCCCGTCCTTGGAAAGCTCCTCGAAAAACTGATCACCATACGGATTATGCATTGGGCATCTGTCACAGGAAGGAGCAGTCAACGACAGTTCGGCTTCAAGGAGCAAACCGGCACGGTCGACGCCATTAACACCCTGATAGACACTATACGAAAAGCCAAACAAGAGAGGTCACAGGTTGTCGCCGTCTCGCTCGACATCAGGGCGGCTTTTGATAACGCCTGGTGGCCGGTCCTATTACACCGCCTTAGAGATATACGATGCCCCAAAAATATCTACGGCCTCATCAAGAGCTACCTCCGAGACAGGCGCGTCGTTCTGGACCACGGAGGAGTTCGCGTCTCCAGAGCCACGTCAAAAGGTTGCGTTCAGGGCTCGGTGTGCGGGCCTGCATTGTGGAACATCATACTTGATGAACTATTGGAGATCCATCTACCATCTGGATGCCACATGCAGGCCTACGCAGACGATGTGGTACTCGTGGTCACCGCCGAAAATGTCAATCGCCTAGAGGAGCTCACCGACACGGTTCTCCAACAGATAGTGAATTGGGGAAAAAGCGTCAAGCTCGAATTCGGGGCCTCCAAAACACAACTGATCGCCTTCACCCCCAAGGCCAGAACTCTTAGGGTGGAGATGGACGGTCAAATCTTAAAAACCTGTAAGGAGATCAAGCTCCTCGGAATAATTTTGGACGAAAAGCTACTCTTTGGCCGCCATGTACAGTACGTCCTGGGAAAAGCCTCGCGAATATTCAACAAACTGTGCTTGTACTCCCGACCAACCTGGGGTGCTCATCCCGAGAACGTGCGCACCATCTATCTCCGAGTCATCGAGCCAATCGTCACCTACGCTGCCGGGGTGTGGGGACACATAGTAAACAAGCGCTACATCAAAAAATCTTTGATGAGCATGCAGCGTGGTTTTGCCCTAAAAGCCATCAGAGGATTCCGCACTATTTCAACCTCCGCTGCTCTGGCTCTCGCACAATTCACCCCTCTAGACCTAAAAATCCGAGAAGTACACCAAATCGAAAAAGTCAGGCTCACTGGGAAAACTGAATTCCTCCCCAATGACATTTCTTATGAGAAACCTGTCCCCGTCAAGGACCTCCTACATCCCGCTAGAAGGCCGACAATCAAACCCATACCCTCCTCTGAACACCGAATTAACTCCGAAATGACGGACAGTACTCTGGTCACTCGCGTATTCACTGACGGAAGTAAGCAGGACGATGGATCAGTTGGAGCCGCATTCGTGCTTTACAGACCCGGCGACACTGAAATATCCGTAACAAAAAAGTACAAATTACACAGTAGCTGCACAGTCTTCCAGGCAGAGCTGTACGCCATCTGGAAAGCCTGTCAGTGGATTGTAAGCGAAAATCATCCCCACACATACATCTTCACCGATTCTCTCTCATCAATATACGCGATAAATAACAGAAGCAACACACACCCCATCATAGTGAACATACACAAACTCATACACGCTAACCACACTACACACAAAATTTTCATAGACTGGGTAAAGGGACACTCGGGTGTCATAGGAAACGAGGAAGCCGATGCGGCTGCCAACAGCGGCGCACGCATGCACAAGGCCCCCGACTACTCGCAGTTCTCAATATCCTACGTGAAACACAAGATTCACACAGAACACCACGCCATCTGGCAAGCCAGATACGAAAGCTCCCCGCAGGGCTCACACACAAAAACACTACTACCAAAACTGAACGACATCAAAGAACTCAACAAATGCACACAAACAAACTTTCAGCTTACACAAATACTCACAGGCCATGGATATCACAAGACATACTTACATCGCTTCAAAATAGCACCAGATGACACCTGCCCCTGCGACGGCACTTCCAGCCAAACTGTAGAACACCTCCTTAAAAACTGCCAAGTGTTCGCTGCGAAAAGGCACAGCCACGAAGAAACGTGCCACGAAGTACAGGTGTCACCATATAATTTACCTGAAATGTTAAAAAAGAAAGCTGCAATCAACTCTTTCACTTCGTTTTGCCAAACAATAATCAACAACATCAAGAAAATAAATGAAAACAATTAG
Protein
MAPVIPEITLPASAEDINSRDTYLETVRKLISCITTAVTEGKSVTAANKRLICSAAEEIRRATRVLETTLASTLTPPTPSPPATNEGLKEEIIACVREEFKRHRQLPPLTLQSQVRPSYAQAAAPNPRKPALTTKPTPKTYPATKPAIIVSAKTEVKTKQDIIEAWRRNISFKNSGFAPSRVQAVSNNKLRIEFDNIEQRNDTLTRITNSQTIHAEPARRLLPMLNLKGVSKDVPAEDLVSIISRQNPELTNAINQKEDLRFRFQRSNRNPNLYNAILIAQPTVWRKCISLGRISVDHQRIHVEEFTPFLQCHKCLSFGHTKNKCSSTEQYCAHCSANNEVKNIPGINIYQFSDGARVKACILAKTGVGSVLGLSQYSSANLSVIQLTAGNKKTYIASTYIEPDVDANGTMERLEVFLKDIGGARVIVGGDFNGWHPSWGSVRVNPRGSSLVELAHSNDLYICNSGDTPTFETITHGRNRSSIIDLTLASSDLFNSISEWHVNLNTCPTSQHNAIEFSLNTNENTNKHAENTSTFLYKSEQANWTIFRNTLHTLMANTDILDRSVESLSPEGLESLLREVTDILHRACRASMRVKAGSGNRHKPPWWNDRLQDLKNEVINVHRRLRAAKRQNLPLDDLIEQQQTKKALYADALRAESTRSFRSFCELQTKENVWSLTNRLLKESTPKRPPATLKTGNTFTTCAKETAEALLEHFYPADTPDTIARHHQLRACYDAPVDTENDLPFTEEEVMECLQYMNPKKAPGLDHITADICQQFASAYPRLITNIMNRCLTLQHFPDQWKTAYVKIIPKPGKSDHTSLTSFRPIGLIPVLGKLLEKLITIRIMHWASVTGRSSQRQFGFKEQTGTVDAINTLIDTIRKAKQERSQVVAVSLDIRAAFDNAWWPVLLHRLRDIRCPKNIYGLIKSYLRDRRVVLDHGGVRVSRATSKGCVQGSVCGPALWNIILDELLEIHLPSGCHMQAYADDVVLVVTAENVNRLEELTDTVLQQIVNWGKSVKLEFGASKTQLIAFTPKARTLRVEMDGQILKTCKEIKLLGIILDEKLLFGRHVQYVLGKASRIFNKLCLYSRPTWGAHPENVRTIYLRVIEPIVTYAAGVWGHIVNKRYIKKSLMSMQRGFALKAIRGFRTISTSAALALAQFTPLDLKIREVHQIEKVRLTGKTEFLPNDISYEKPVPVKDLLHPARRPTIKPIPSSEHRINSEMTDSTLVTRVFTDGSKQDDGSVGAAFVLYRPGDTEISVTKKYKLHSSCTVFQAELYAIWKACQWIVSENHPHTYIFTDSLSSIYAINNRSNTHPIIVNIHKLIHANHTTHKIFIDWVKGHSGVIGNEEADAAANSGARMHKAPDYSQFSISYVKHKIHTEHHAIWQARYESSPQGSHTKTLLPKLNDIKELNKCTQTNFQLTQILTGHGYHKTYLHRFKIAPDDTCPCDGTSSQTVEHLLKNCQVFAAKRHSHEETCHEVQVSPYNLPEMLKKKAAINSFTSFCQTIINNIKKINENN
Summary
Uniprot
Q93139
Q9BLI5
A0A1W7R9Y4
A0A224X5M4
A0A0K1IK29
A0A023EX80
+ More
A0A0K1IK14 A0A087TAD8 A0A087TI44 A0A087UHT1 A0A1B6G0A1 A0A142LX31 A0A087TUG6 U5EVA2 K7JMI9 A0A142LX39 A0A142LX24 U5EV73 U5EW32 A0A224X5X2 O18558 U5EPW8 A0A0G2LA86 A0A142LX33 A0A226DPQ5 J9MA04 A0A0G2KM65 U5EG91 U5ERW4 A0A226DQD5 A0A226E8J9 K7JA12 U5ES21
A0A0K1IK14 A0A087TAD8 A0A087TI44 A0A087UHT1 A0A1B6G0A1 A0A142LX31 A0A087TUG6 U5EVA2 K7JMI9 A0A142LX39 A0A142LX24 U5EV73 U5EW32 A0A224X5X2 O18558 U5EPW8 A0A0G2LA86 A0A142LX33 A0A226DPQ5 J9MA04 A0A0G2KM65 U5EG91 U5ERW4 A0A226DQD5 A0A226E8J9 K7JA12 U5ES21
EMBL
D38414
BAA07467.1
AB046668
BAB21511.1
GFAH01000444
JAV47945.1
+ More
GFTR01008639 JAW07787.1 KP771711 AKU04648.1 GBBI01004920 JAC13792.1 KP771712 AKU04650.1 KK114278 KFM62077.1 KK115320 KFM64783.1 KK119855 KFM76920.1 GECZ01013919 JAS55850.1 KU543676 AMS38357.1 KK116791 KFM68755.1 GANO01003496 JAB56375.1 AAZX01008070 KU543680 AMS38365.1 KU543672 AMS38350.1 GANO01003537 JAB56334.1 GANO01003141 JAB56730.1 GFTR01008541 JAW07885.1 U87543 AAB65093.1 GANO01003536 JAB56335.1 CR848788 KU543677 AMS38359.1 LNIX01000015 OXA46641.1 ABLF02008888 ABLF02016960 ABLF02032572 BX908731 GANO01003539 JAB56332.1 GANO01003538 JAB56333.1 LNIX01000013 OXA47423.1 LNIX01000005 OXA53875.1 AAZX01016607 AAZX01023407 GANO01003451 JAB56420.1
GFTR01008639 JAW07787.1 KP771711 AKU04648.1 GBBI01004920 JAC13792.1 KP771712 AKU04650.1 KK114278 KFM62077.1 KK115320 KFM64783.1 KK119855 KFM76920.1 GECZ01013919 JAS55850.1 KU543676 AMS38357.1 KK116791 KFM68755.1 GANO01003496 JAB56375.1 AAZX01008070 KU543680 AMS38365.1 KU543672 AMS38350.1 GANO01003537 JAB56334.1 GANO01003141 JAB56730.1 GFTR01008541 JAW07885.1 U87543 AAB65093.1 GANO01003536 JAB56335.1 CR848788 KU543677 AMS38359.1 LNIX01000015 OXA46641.1 ABLF02008888 ABLF02016960 ABLF02032572 BX908731 GANO01003539 JAB56332.1 GANO01003538 JAB56333.1 LNIX01000013 OXA47423.1 LNIX01000005 OXA53875.1 AAZX01016607 AAZX01023407 GANO01003451 JAB56420.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
Q93139
Q9BLI5
A0A1W7R9Y4
A0A224X5M4
A0A0K1IK29
A0A023EX80
+ More
A0A0K1IK14 A0A087TAD8 A0A087TI44 A0A087UHT1 A0A1B6G0A1 A0A142LX31 A0A087TUG6 U5EVA2 K7JMI9 A0A142LX39 A0A142LX24 U5EV73 U5EW32 A0A224X5X2 O18558 U5EPW8 A0A0G2LA86 A0A142LX33 A0A226DPQ5 J9MA04 A0A0G2KM65 U5EG91 U5ERW4 A0A226DQD5 A0A226E8J9 K7JA12 U5ES21
A0A0K1IK14 A0A087TAD8 A0A087TI44 A0A087UHT1 A0A1B6G0A1 A0A142LX31 A0A087TUG6 U5EVA2 K7JMI9 A0A142LX39 A0A142LX24 U5EV73 U5EW32 A0A224X5X2 O18558 U5EPW8 A0A0G2LA86 A0A142LX33 A0A226DPQ5 J9MA04 A0A0G2KM65 U5EG91 U5ERW4 A0A226DQD5 A0A226E8J9 K7JA12 U5ES21
PDB
1WDU
E-value=8.36275e-16,
Score=210
Ontologies
KEGG
GO
PANTHER
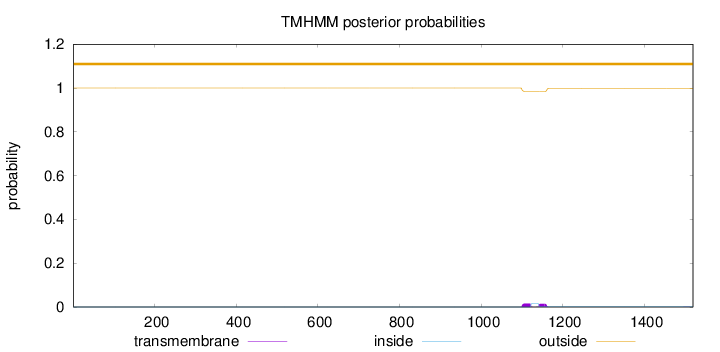
Topology
Length:
1519
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.61052
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00003
outside
1 - 1519
Population Genetic Test Statistics
Pi
233.226927
Theta
17.749699
Tajima's D
0.481843
CLR
13827.414143
CSRT
0.51812409379531
Interpretation
Uncertain
