Gene
KWMTBOMO07918
Pre Gene Modal
BGIBMGA001173
Annotation
PREDICTED:_alkaline_nuclease_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.405
Sequence
CDS
ATGCGTCTGACGCTTGTTCTCGCAGCATTAGCTGTAGCGGTGGTGGCACTGCCCTCCAAACTTCGAACGGATCTACCAGAGCCAGGACAACTGGCTTTTGTTCTTAACGAAGATGAGTTTGAAGACTACCTCGATGCCTATCTCGCTCTTCAACAATCTGAAATGCTCGCCAATCAAACCAGGAATGATTTCCGAAGCGGTTGCACATTCCGAGTCAACGGTGATCTTGGTCAGCCTCAACCTGTATATATCCACAGAGGAAATTATTTAAGTCCAACTGGTAATACAGGTCAGATTCGTCTTAATCGTGGGGAACAAGTTCTTATCGCTTGCACAGGATCCGGCCGTACGATCCGTCACCCCAATGTTGCTTCTAACCTTGCTGTCGGGACGGTCAGCTGTCAAAACAATAATCTCGTCACAGCCAATTGGCTGCGTGGTAACAGTGCTTTCGGGCAACTCACATGCTCTTCTCACGCTCACCATGACGCTCAACAAACCAACACTAGATGCTTCAATAATCACTTTGTTATTCGTGTGGGATTCATCGTCAACAATGTATTTTATCCTCTATATTGGTCCTGCTTTGACCGCAACCGCTTAGAGGTTCTATATGTATGGTACCACCAAAACCCACCTAATTCCGTATTCCAGTCTCGCGTTGATAGGCCTAGTTGGATAGCTGGAAACTTCTTCCCCGGTGTAGCTGTTAACTCTGCCTATACTCAAGTATCTCAAAGAAACATGATCGCTGGATTCGTGGGTAATGCCCTCGCAGACAGATATGTAACGTCCACGCAGTTCTTAGCACGGGGTCATCTTGCTGCTAAGACTGACTTTATATATGCAACCGGTCAACGTGCTAGCTTCTACTTTATTAACGCTGCTCCTCAATGGCAGCCTTTCAACGCTGGAAACTGGAACCGGCTCGAGCAGAATCTTCGTCGTAGAATTGGACAGGCTGGATATCATACCATGGTCTACACTGGAACATTCCGCGTAACACAACTTAGAAATCAAAACAACCGACTTGTTGATATTTTCTTGCATCGTGCATCCAACGGCGCTCTTCAAATTCCTGTACCACTTTATTTCTACAAGGTGGTGCATGACTCCTCTCGTCGCCTCGGTACCGCCTTTATCAGTATCAACAATCCTTACTACACCCAAGCGGAGGCTCGCAATCTGCAGTTCTGCACAGACCGTTGCCGCAATAACAACGCATTCAACTGGGTGGGGTGGCAACCTGATCGCATCGACCTCGGCTATAGCTTCTGCTGTACCATTGCTGATTTCCGAAGAACTATTCCGCACCTTCCAGCTTTCAATGTTAACGGACTTCTGACATAA
Protein
MRLTLVLAALAVAVVALPSKLRTDLPEPGQLAFVLNEDEFEDYLDAYLALQQSEMLANQTRNDFRSGCTFRVNGDLGQPQPVYIHRGNYLSPTGNTGQIRLNRGEQVLIACTGSGRTIRHPNVASNLAVGTVSCQNNNLVTANWLRGNSAFGQLTCSSHAHHDAQQTNTRCFNNHFVIRVGFIVNNVFYPLYWSCFDRNRLEVLYVWYHQNPPNSVFQSRVDRPSWIAGNFFPGVAVNSAYTQVSQRNMIAGFVGNALADRYVTSTQFLARGHLAAKTDFIYATGQRASFYFINAAPQWQPFNAGNWNRLEQNLRRRIGQAGYHTMVYTGTFRVTQLRNQNNRLVDIFLHRASNGALQIPVPLYFYKVVHDSSRRLGTAFISINNPYYTQAEARNLQFCTDRCRNNNAFNWVGWQPDRIDLGYSFCCTIADFRRTIPHLPAFNVNGLLT
Summary
Uniprot
Q08JX1
A0A0N1IIN7
A0A194Q8S9
A0A2W1BYI3
A0A0N1IB53
A0A2A4JP76
+ More
A0A2H1WP54 A0A219LVV6 A0A2A4JML0 F6K708 A0A2H1W493 B7VCC2 A0A194Q8R2 B7VCC3 A0A1E1WMF9 A0A219LVU7 A0A2A4JNR7 A0A2W1BP50 A0A194RKW6 A0A2H1V3T5 A0A219LVV5 A0A219LVV4 A0A0L7KR40 A0A1E1WTA0 A0A2H1WT12 A0A0N1ICG2 A0A2A4JKE1 A0A2W1BV74 A0A0L7LLE8 H9JIU6 A0A219LWI3 A0A0N1PH39 A0A194Q7A2 A0A0N1II16 A0A1B0CE89 A0A1L8DXI7 A0A3S2NR20 A0A0N0PDC8 A0A1L8DPN4 A0A1L8DXM9 B0WU96 A0A2P8ZMT4 A0A1J1IDA7 A0A2H1VP74 A0A1S4FDC9 Q176L0 A0A182RUK6 A0A336LKP8 W5JF80 A0A182M5I5 A0A182G849 W5JEZ9 A0A182QZC1 A0A182M6U5 A0A182WDU5 A0A212EIS2 A0A067RF87 A0A182VCF7 W8BUZ2 A0A182HW10 A0A182YBI8 W8BEM3 A0A182QLU4 A7UR43 A0A182JWX9 A0A182J459 A0A3B0JT56 A0A182FVL7 A0A0A1WJV0 A0A182WUG9 A0A182IU94 A0A1W4UQR3 A0A034WII0 A0A1A9UG98 B4H6U2 A0A182WQS2 Q29DJ9 B4IFY0 A0A1L8EG88 A0A0J9RZ63 A0A1L8EG81 A0A1L8EGD9 A0A1B0C252 A0A1A9ZRY2 A0A182PHB8 B3M7Q7 B4N4S4 A0A182N3S5 A0A182HW09 A0A219LVW0 A0A1I8NNQ2 A0A182WUG8 B4PH14 A0A1A9YID0
A0A2H1WP54 A0A219LVV6 A0A2A4JML0 F6K708 A0A2H1W493 B7VCC2 A0A194Q8R2 B7VCC3 A0A1E1WMF9 A0A219LVU7 A0A2A4JNR7 A0A2W1BP50 A0A194RKW6 A0A2H1V3T5 A0A219LVV5 A0A219LVV4 A0A0L7KR40 A0A1E1WTA0 A0A2H1WT12 A0A0N1ICG2 A0A2A4JKE1 A0A2W1BV74 A0A0L7LLE8 H9JIU6 A0A219LWI3 A0A0N1PH39 A0A194Q7A2 A0A0N1II16 A0A1B0CE89 A0A1L8DXI7 A0A3S2NR20 A0A0N0PDC8 A0A1L8DPN4 A0A1L8DXM9 B0WU96 A0A2P8ZMT4 A0A1J1IDA7 A0A2H1VP74 A0A1S4FDC9 Q176L0 A0A182RUK6 A0A336LKP8 W5JF80 A0A182M5I5 A0A182G849 W5JEZ9 A0A182QZC1 A0A182M6U5 A0A182WDU5 A0A212EIS2 A0A067RF87 A0A182VCF7 W8BUZ2 A0A182HW10 A0A182YBI8 W8BEM3 A0A182QLU4 A7UR43 A0A182JWX9 A0A182J459 A0A3B0JT56 A0A182FVL7 A0A0A1WJV0 A0A182WUG9 A0A182IU94 A0A1W4UQR3 A0A034WII0 A0A1A9UG98 B4H6U2 A0A182WQS2 Q29DJ9 B4IFY0 A0A1L8EG88 A0A0J9RZ63 A0A1L8EG81 A0A1L8EGD9 A0A1B0C252 A0A1A9ZRY2 A0A182PHB8 B3M7Q7 B4N4S4 A0A182N3S5 A0A182HW09 A0A219LVW0 A0A1I8NNQ2 A0A182WUG8 B4PH14 A0A1A9YID0
Pubmed
EMBL
AB254196
BAF33251.1
KQ460202
KPJ17191.1
KQ459299
KPJ01928.1
+ More
KZ149944 PZC76833.1 KPJ17193.1 NWSH01000988 PCG73222.1 ODYU01009979 SOQ54756.1 KM229550 AKB95589.1 PCG73221.1 HM357845 AEA76311.1 ODYU01006163 SOQ47776.1 FM253095 CAR92521.1 KPJ01927.1 FM253096 CAR92522.1 GDQN01002889 JAT88165.1 KM229544 AKB95583.1 PCG73224.1 PZC76832.1 KQ460045 KPJ18059.1 ODYU01000364 SOQ35022.1 KM229545 AKB95584.1 KM229551 AKB95590.1 JTDY01007018 KOB65521.1 GDQN01000872 JAT90182.1 ODYU01010450 SOQ55574.1 KQ458751 KPJ05311.1 NWSH01001168 PCG72289.1 KZ149927 PZC77534.1 JTDY01000675 KOB76267.1 BABH01023166 KM229547 AKB95586.1 KQ460396 KPJ15353.1 KQ459580 KPI99275.1 KQ459986 KPJ18843.1 AJWK01008671 GFDF01002907 JAV11177.1 RSAL01000132 RVE46356.1 KPJ15345.1 GFDF01005662 JAV08422.1 GFDF01002908 JAV11176.1 DS232102 EDS34867.1 PYGN01000014 PSN57821.1 CVRI01000047 CRK98271.1 ODYU01003633 SOQ42630.1 CH477386 EAT42072.1 UFQT01000005 SSX17321.1 ADMH02001680 ETN61459.1 AXCM01003118 JXUM01047237 KQ561511 KXJ78332.1 ETN61460.1 AXCN02001098 AGBW02014574 OWR41393.1 KK852672 KDR18788.1 GAMC01009439 JAB97116.1 APCN01001456 GAMC01009438 JAB97117.1 AAAB01008804 EDO64808.1 OUUW01000002 SPP76536.1 GBXI01015003 JAC99288.1 GAKP01004820 JAC54132.1 CH479215 EDW33530.1 CH379070 EAL30415.1 CH480834 EDW46567.1 GFDG01001154 JAV17645.1 CM002912 KMZ00475.1 GFDG01001153 JAV17646.1 GFDG01001156 JAV17643.1 JXJN01024350 CH902618 EDV40985.1 CH964095 EDW79148.1 KM229549 AKB95588.1 CM000159 EDW95384.1
KZ149944 PZC76833.1 KPJ17193.1 NWSH01000988 PCG73222.1 ODYU01009979 SOQ54756.1 KM229550 AKB95589.1 PCG73221.1 HM357845 AEA76311.1 ODYU01006163 SOQ47776.1 FM253095 CAR92521.1 KPJ01927.1 FM253096 CAR92522.1 GDQN01002889 JAT88165.1 KM229544 AKB95583.1 PCG73224.1 PZC76832.1 KQ460045 KPJ18059.1 ODYU01000364 SOQ35022.1 KM229545 AKB95584.1 KM229551 AKB95590.1 JTDY01007018 KOB65521.1 GDQN01000872 JAT90182.1 ODYU01010450 SOQ55574.1 KQ458751 KPJ05311.1 NWSH01001168 PCG72289.1 KZ149927 PZC77534.1 JTDY01000675 KOB76267.1 BABH01023166 KM229547 AKB95586.1 KQ460396 KPJ15353.1 KQ459580 KPI99275.1 KQ459986 KPJ18843.1 AJWK01008671 GFDF01002907 JAV11177.1 RSAL01000132 RVE46356.1 KPJ15345.1 GFDF01005662 JAV08422.1 GFDF01002908 JAV11176.1 DS232102 EDS34867.1 PYGN01000014 PSN57821.1 CVRI01000047 CRK98271.1 ODYU01003633 SOQ42630.1 CH477386 EAT42072.1 UFQT01000005 SSX17321.1 ADMH02001680 ETN61459.1 AXCM01003118 JXUM01047237 KQ561511 KXJ78332.1 ETN61460.1 AXCN02001098 AGBW02014574 OWR41393.1 KK852672 KDR18788.1 GAMC01009439 JAB97116.1 APCN01001456 GAMC01009438 JAB97117.1 AAAB01008804 EDO64808.1 OUUW01000002 SPP76536.1 GBXI01015003 JAC99288.1 GAKP01004820 JAC54132.1 CH479215 EDW33530.1 CH379070 EAL30415.1 CH480834 EDW46567.1 GFDG01001154 JAV17645.1 CM002912 KMZ00475.1 GFDG01001153 JAV17646.1 GFDG01001156 JAV17643.1 JXJN01024350 CH902618 EDV40985.1 CH964095 EDW79148.1 KM229549 AKB95588.1 CM000159 EDW95384.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000037510
UP000005204
UP000092461
+ More
UP000283053 UP000002320 UP000245037 UP000183832 UP000008820 UP000075900 UP000000673 UP000075883 UP000069940 UP000249989 UP000075886 UP000075920 UP000007151 UP000027135 UP000075903 UP000075840 UP000076408 UP000007062 UP000075881 UP000075880 UP000268350 UP000069272 UP000076407 UP000192221 UP000078200 UP000008744 UP000001819 UP000001292 UP000092460 UP000092445 UP000075885 UP000007801 UP000007798 UP000075884 UP000095300 UP000002282 UP000092443
UP000283053 UP000002320 UP000245037 UP000183832 UP000008820 UP000075900 UP000000673 UP000075883 UP000069940 UP000249989 UP000075886 UP000075920 UP000007151 UP000027135 UP000075903 UP000075840 UP000076408 UP000007062 UP000075881 UP000075880 UP000268350 UP000069272 UP000076407 UP000192221 UP000078200 UP000008744 UP000001819 UP000001292 UP000092460 UP000092445 UP000075885 UP000007801 UP000007798 UP000075884 UP000095300 UP000002282 UP000092443
Interpro
IPR001604
DNA/RNA_non-sp_Endonuclease
+ More
IPR040255 Non-specific_endonuclease
IPR020821 Extracellular_endonuc_su_A
IPR015324 Ribosomal_Rsm22-like
IPR029063 SAM-dependent_MTases
IPR038518 Glyco_hydro_63N_sf
IPR031631 Glyco_hydro_63N
IPR008928 6-hairpin_glycosidase_sf
IPR012341 6hp_glycosidase-like_sf
IPR031335 Glyco_hydro_63_C
IPR004888 Glycoside_hydrolase_63
IPR040255 Non-specific_endonuclease
IPR020821 Extracellular_endonuc_su_A
IPR015324 Ribosomal_Rsm22-like
IPR029063 SAM-dependent_MTases
IPR038518 Glyco_hydro_63N_sf
IPR031631 Glyco_hydro_63N
IPR008928 6-hairpin_glycosidase_sf
IPR012341 6hp_glycosidase-like_sf
IPR031335 Glyco_hydro_63_C
IPR004888 Glycoside_hydrolase_63
Gene 3D
ProteinModelPortal
Q08JX1
A0A0N1IIN7
A0A194Q8S9
A0A2W1BYI3
A0A0N1IB53
A0A2A4JP76
+ More
A0A2H1WP54 A0A219LVV6 A0A2A4JML0 F6K708 A0A2H1W493 B7VCC2 A0A194Q8R2 B7VCC3 A0A1E1WMF9 A0A219LVU7 A0A2A4JNR7 A0A2W1BP50 A0A194RKW6 A0A2H1V3T5 A0A219LVV5 A0A219LVV4 A0A0L7KR40 A0A1E1WTA0 A0A2H1WT12 A0A0N1ICG2 A0A2A4JKE1 A0A2W1BV74 A0A0L7LLE8 H9JIU6 A0A219LWI3 A0A0N1PH39 A0A194Q7A2 A0A0N1II16 A0A1B0CE89 A0A1L8DXI7 A0A3S2NR20 A0A0N0PDC8 A0A1L8DPN4 A0A1L8DXM9 B0WU96 A0A2P8ZMT4 A0A1J1IDA7 A0A2H1VP74 A0A1S4FDC9 Q176L0 A0A182RUK6 A0A336LKP8 W5JF80 A0A182M5I5 A0A182G849 W5JEZ9 A0A182QZC1 A0A182M6U5 A0A182WDU5 A0A212EIS2 A0A067RF87 A0A182VCF7 W8BUZ2 A0A182HW10 A0A182YBI8 W8BEM3 A0A182QLU4 A7UR43 A0A182JWX9 A0A182J459 A0A3B0JT56 A0A182FVL7 A0A0A1WJV0 A0A182WUG9 A0A182IU94 A0A1W4UQR3 A0A034WII0 A0A1A9UG98 B4H6U2 A0A182WQS2 Q29DJ9 B4IFY0 A0A1L8EG88 A0A0J9RZ63 A0A1L8EG81 A0A1L8EGD9 A0A1B0C252 A0A1A9ZRY2 A0A182PHB8 B3M7Q7 B4N4S4 A0A182N3S5 A0A182HW09 A0A219LVW0 A0A1I8NNQ2 A0A182WUG8 B4PH14 A0A1A9YID0
A0A2H1WP54 A0A219LVV6 A0A2A4JML0 F6K708 A0A2H1W493 B7VCC2 A0A194Q8R2 B7VCC3 A0A1E1WMF9 A0A219LVU7 A0A2A4JNR7 A0A2W1BP50 A0A194RKW6 A0A2H1V3T5 A0A219LVV5 A0A219LVV4 A0A0L7KR40 A0A1E1WTA0 A0A2H1WT12 A0A0N1ICG2 A0A2A4JKE1 A0A2W1BV74 A0A0L7LLE8 H9JIU6 A0A219LWI3 A0A0N1PH39 A0A194Q7A2 A0A0N1II16 A0A1B0CE89 A0A1L8DXI7 A0A3S2NR20 A0A0N0PDC8 A0A1L8DPN4 A0A1L8DXM9 B0WU96 A0A2P8ZMT4 A0A1J1IDA7 A0A2H1VP74 A0A1S4FDC9 Q176L0 A0A182RUK6 A0A336LKP8 W5JF80 A0A182M5I5 A0A182G849 W5JEZ9 A0A182QZC1 A0A182M6U5 A0A182WDU5 A0A212EIS2 A0A067RF87 A0A182VCF7 W8BUZ2 A0A182HW10 A0A182YBI8 W8BEM3 A0A182QLU4 A7UR43 A0A182JWX9 A0A182J459 A0A3B0JT56 A0A182FVL7 A0A0A1WJV0 A0A182WUG9 A0A182IU94 A0A1W4UQR3 A0A034WII0 A0A1A9UG98 B4H6U2 A0A182WQS2 Q29DJ9 B4IFY0 A0A1L8EG88 A0A0J9RZ63 A0A1L8EG81 A0A1L8EGD9 A0A1B0C252 A0A1A9ZRY2 A0A182PHB8 B3M7Q7 B4N4S4 A0A182N3S5 A0A182HW09 A0A219LVW0 A0A1I8NNQ2 A0A182WUG8 B4PH14 A0A1A9YID0
PDB
4A1N
E-value=0.00713373,
Score=93
Ontologies
GO
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.990527
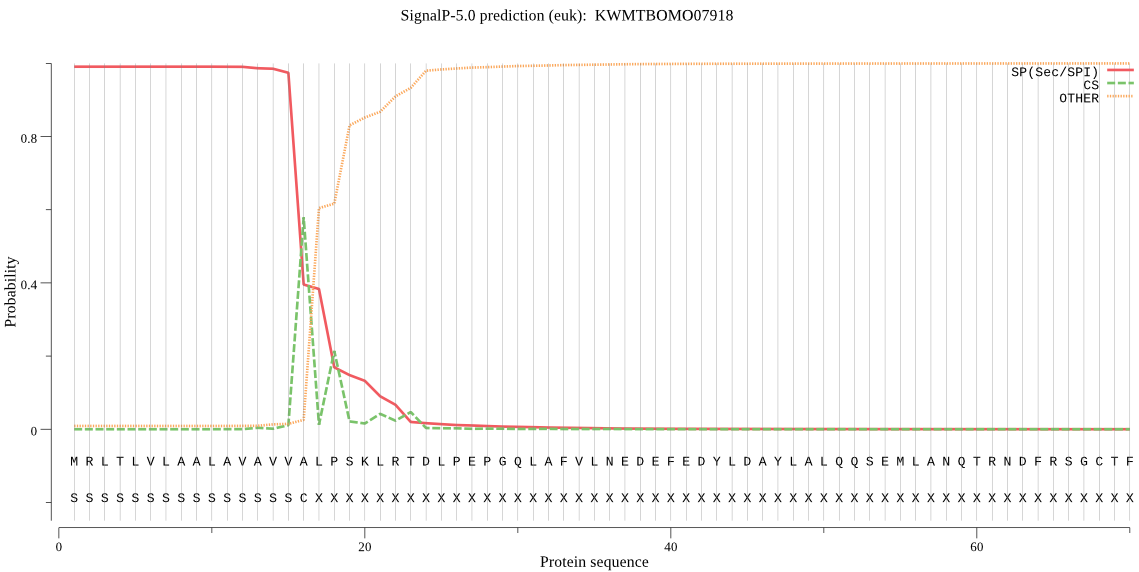
Length:
449
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.67706
Exp number, first 60 AAs:
0.41892
Total prob of N-in:
0.07243
outside
1 - 449

Population Genetic Test Statistics
Pi
80.240386
Theta
112.90211
Tajima's D
-0.810127
CLR
433.290654
CSRT
0.179491025448728
Interpretation
Uncertain
