Gene
KWMTBOMO07915
Annotation
PREDICTED:_alkaline_nuclease_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.178
Sequence
CDS
ATGGAAAAAGTAAAACAGACTTCGGAAGCAACAAAAACATCAAAACTCCGTGCACGCATTGGTGAAGCAGGATATAATACTACTGTTTACACGGGAACCTTCGGCGTAACCCAACTACGGGACCAAAATGGCCAACTTGTTGATATTTACCTTTACCGGGACACTAACAATAACCCTCAGATACCTGTTCCGTTGTACTTTTACAAAGTTGTGTATGAGCCTGTCCTACAAAAGGGCACAGCTTTTGTGGCGATAAACAACCCGTACTACACTGAAACTGAAGTACGCGCCTTGCAATTCTGCACTGACCACTGCCGCAACAACACCGCGTTCAACTGGATTGGCTGGCAACCAGATCGAATCGACATCGGGTACAGCTTCTGCTGTACCATCGAAGACTTTAGACGGGCTGTGCCGCATTTGCCAGACTTTAATGTTACCGGGCTTTTGTACTGA
Protein
MEKVKQTSEATKTSKLRARIGEAGYNTTVYTGTFGVTQLRDQNGQLVDIYLYRDTNNNPQIPVPLYFYKVVYEPVLQKGTAFVAINNPYYTETEVRALQFCTDHCRNNTAFNWIGWQPDRIDIGYSFCCTIEDFRRAVPHLPDFNVTGLLY
Summary
Uniprot
A0A2H1V3T5
A0A1E1WMF9
A0A219LVV4
H9IV93
A0A2A4JNR7
A0A2W1BP50
+ More
Q08JX1 A0A2H1WP54 A0A2A4JML0 B7VCC3 A0A219LVV6 A0A2A4JP76 A0A194Q8Y6 A0A2W1BYI3 A0A1E1WTA0 A0A194RKW6 B7VCC2 F6K708 A0A2H1W493 A0A219LVU7 A0A219LVV5 A0A0N1IB53 A0A194Q8R2 A0A194Q8S9 A0A0N1IIN7 A0A212ETI7 A0A0N1ICG2 A0A212FE21 A0A2W1BV74 A0A0L7LLE8 A0A2A4JKE1 A0A219LWI3 H9JIU6 A0A0L7KR40 A0A2H1WT12 A0A0N1PH39 A0A3S2NR20 A0A219LVU2 A0A194Q7A2 A0A2A4K9C7 A0A0N0PDC8 A0A212FJT7 A0A2H1VP74 A0A2H1W9B5 B4N4S4 B4PH14 A0A0J9RZ63 B4QQB0 Q9VVV1 B3NDV9 B3M7Q7 A0A1B0CE89 A0A212EIS2 B4IFY0 T1P9F7 W8BUZ2 W8BEM3 A0A1W4UQR3 A0A1W4XR54 A0A1I8MCL2 A0A2P8ZMT4 A0A067RF87 A0A0P5K0P6 A0A0N8B5M4 A0A0P5W6R4 A0A0P5TF11 A0A0P6C109 A0A3B0JT56 A0A0M4EDJ3 B4IYD0 A0A1L8DXM9 A0A1L8DXI7 A0A1L8DPN4 A0A164USM9 Q29DJ9 B4H6U2 A0A1B0D5G0 A0A1A9YID0 A0A1B0C252 B4N4S6 A0A1A9UG98 D3TPG4 A0A0P5FLY5 A0A1B0FEL1 B4KZ79 Q29DK1 B4H6U4 A0A0P5L6U2 A0A0P5JV63 A0A0P5RYT1 A0A0P5S977 A0A0P5NE77 A0A0P5S216 A0A0N8CAS7 A0A0P5NWM0 A0A0P5NP42 A0A0P5S6M9
Q08JX1 A0A2H1WP54 A0A2A4JML0 B7VCC3 A0A219LVV6 A0A2A4JP76 A0A194Q8Y6 A0A2W1BYI3 A0A1E1WTA0 A0A194RKW6 B7VCC2 F6K708 A0A2H1W493 A0A219LVU7 A0A219LVV5 A0A0N1IB53 A0A194Q8R2 A0A194Q8S9 A0A0N1IIN7 A0A212ETI7 A0A0N1ICG2 A0A212FE21 A0A2W1BV74 A0A0L7LLE8 A0A2A4JKE1 A0A219LWI3 H9JIU6 A0A0L7KR40 A0A2H1WT12 A0A0N1PH39 A0A3S2NR20 A0A219LVU2 A0A194Q7A2 A0A2A4K9C7 A0A0N0PDC8 A0A212FJT7 A0A2H1VP74 A0A2H1W9B5 B4N4S4 B4PH14 A0A0J9RZ63 B4QQB0 Q9VVV1 B3NDV9 B3M7Q7 A0A1B0CE89 A0A212EIS2 B4IFY0 T1P9F7 W8BUZ2 W8BEM3 A0A1W4UQR3 A0A1W4XR54 A0A1I8MCL2 A0A2P8ZMT4 A0A067RF87 A0A0P5K0P6 A0A0N8B5M4 A0A0P5W6R4 A0A0P5TF11 A0A0P6C109 A0A3B0JT56 A0A0M4EDJ3 B4IYD0 A0A1L8DXM9 A0A1L8DXI7 A0A1L8DPN4 A0A164USM9 Q29DJ9 B4H6U2 A0A1B0D5G0 A0A1A9YID0 A0A1B0C252 B4N4S6 A0A1A9UG98 D3TPG4 A0A0P5FLY5 A0A1B0FEL1 B4KZ79 Q29DK1 B4H6U4 A0A0P5L6U2 A0A0P5JV63 A0A0P5RYT1 A0A0P5S977 A0A0P5NE77 A0A0P5S216 A0A0N8CAS7 A0A0P5NWM0 A0A0P5NP42 A0A0P5S6M9
Pubmed
EMBL
ODYU01000364
SOQ35022.1
GDQN01002889
JAT88165.1
KM229551
AKB95590.1
+ More
BABH01000729 NWSH01000988 PCG73224.1 KZ149944 PZC76832.1 AB254196 BAF33251.1 ODYU01009979 SOQ54756.1 PCG73221.1 FM253096 CAR92522.1 KM229550 AKB95589.1 PCG73222.1 KQ459324 KPJ01440.1 PZC76833.1 GDQN01000872 JAT90182.1 KQ460045 KPJ18059.1 FM253095 CAR92521.1 HM357845 AEA76311.1 ODYU01006163 SOQ47776.1 KM229544 AKB95583.1 KM229545 AKB95584.1 KQ460202 KPJ17193.1 KQ459299 KPJ01927.1 KPJ01928.1 KPJ17191.1 AGBW02012569 OWR44806.1 KQ458751 KPJ05311.1 AGBW02008987 OWR51970.1 KZ149927 PZC77534.1 JTDY01000675 KOB76267.1 NWSH01001168 PCG72289.1 KM229547 AKB95586.1 BABH01023166 JTDY01007018 KOB65521.1 ODYU01010450 SOQ55574.1 KQ460396 KPJ15353.1 RSAL01000132 RVE46356.1 KM229548 AKB95587.1 KQ459580 KPI99275.1 NWSH01000015 PCG80821.1 KPJ15345.1 AGBW02008242 OWR53970.1 ODYU01003633 SOQ42630.1 ODYU01007039 SOQ49432.1 CH964095 EDW79148.1 CM000159 EDW95384.1 CM002912 KMZ00475.1 CM000363 EDX11016.1 AE014296 AY113510 AAF49206.1 AAM29515.1 CH954178 EDV52313.1 CH902618 EDV40985.1 AJWK01008671 AGBW02014574 OWR41393.1 CH480834 EDW46567.1 KA645411 AFP60040.1 GAMC01009439 JAB97116.1 GAMC01009438 JAB97117.1 PYGN01000014 PSN57821.1 KK852672 KDR18788.1 GDIQ01191520 JAK60205.1 GDIQ01210467 GDIP01099360 JAK41258.1 JAM04355.1 GDIP01090536 JAM13179.1 GDIP01127749 JAL75965.1 GDIP01022692 JAM81023.1 OUUW01000002 SPP76536.1 CP012525 ALC43684.1 CH916366 EDV97603.1 GFDF01002908 JAV11176.1 GFDF01002907 JAV11177.1 GFDF01005662 JAV08422.1 LRGB01001574 KZS11632.1 CH379070 EAL30415.1 CH479215 EDW33530.1 AJVK01011750 AJVK01011751 AJVK01011752 JXJN01024350 EDW79150.1 EZ423316 ADD19592.1 GDIQ01255922 JAJ95802.1 CCAG010013884 CH933809 EDW18905.1 EAL30413.1 EDW33532.1 GDIQ01199212 JAK52513.1 GDIQ01199210 JAK52515.1 GDIQ01099449 JAL52277.1 GDIQ01099446 JAL52280.1 GDIQ01143782 JAL07944.1 GDIQ01098041 JAL53685.1 GDIQ01098043 JAL53683.1 GDIQ01143783 JAL07943.1 GDIQ01143785 JAL07941.1 GDIQ01098044 JAL53682.1
BABH01000729 NWSH01000988 PCG73224.1 KZ149944 PZC76832.1 AB254196 BAF33251.1 ODYU01009979 SOQ54756.1 PCG73221.1 FM253096 CAR92522.1 KM229550 AKB95589.1 PCG73222.1 KQ459324 KPJ01440.1 PZC76833.1 GDQN01000872 JAT90182.1 KQ460045 KPJ18059.1 FM253095 CAR92521.1 HM357845 AEA76311.1 ODYU01006163 SOQ47776.1 KM229544 AKB95583.1 KM229545 AKB95584.1 KQ460202 KPJ17193.1 KQ459299 KPJ01927.1 KPJ01928.1 KPJ17191.1 AGBW02012569 OWR44806.1 KQ458751 KPJ05311.1 AGBW02008987 OWR51970.1 KZ149927 PZC77534.1 JTDY01000675 KOB76267.1 NWSH01001168 PCG72289.1 KM229547 AKB95586.1 BABH01023166 JTDY01007018 KOB65521.1 ODYU01010450 SOQ55574.1 KQ460396 KPJ15353.1 RSAL01000132 RVE46356.1 KM229548 AKB95587.1 KQ459580 KPI99275.1 NWSH01000015 PCG80821.1 KPJ15345.1 AGBW02008242 OWR53970.1 ODYU01003633 SOQ42630.1 ODYU01007039 SOQ49432.1 CH964095 EDW79148.1 CM000159 EDW95384.1 CM002912 KMZ00475.1 CM000363 EDX11016.1 AE014296 AY113510 AAF49206.1 AAM29515.1 CH954178 EDV52313.1 CH902618 EDV40985.1 AJWK01008671 AGBW02014574 OWR41393.1 CH480834 EDW46567.1 KA645411 AFP60040.1 GAMC01009439 JAB97116.1 GAMC01009438 JAB97117.1 PYGN01000014 PSN57821.1 KK852672 KDR18788.1 GDIQ01191520 JAK60205.1 GDIQ01210467 GDIP01099360 JAK41258.1 JAM04355.1 GDIP01090536 JAM13179.1 GDIP01127749 JAL75965.1 GDIP01022692 JAM81023.1 OUUW01000002 SPP76536.1 CP012525 ALC43684.1 CH916366 EDV97603.1 GFDF01002908 JAV11176.1 GFDF01002907 JAV11177.1 GFDF01005662 JAV08422.1 LRGB01001574 KZS11632.1 CH379070 EAL30415.1 CH479215 EDW33530.1 AJVK01011750 AJVK01011751 AJVK01011752 JXJN01024350 EDW79150.1 EZ423316 ADD19592.1 GDIQ01255922 JAJ95802.1 CCAG010013884 CH933809 EDW18905.1 EAL30413.1 EDW33532.1 GDIQ01199212 JAK52513.1 GDIQ01199210 JAK52515.1 GDIQ01099449 JAL52277.1 GDIQ01099446 JAL52280.1 GDIQ01143782 JAL07944.1 GDIQ01098041 JAL53685.1 GDIQ01098043 JAL53683.1 GDIQ01143783 JAL07943.1 GDIQ01143785 JAL07941.1 GDIQ01098044 JAL53682.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000283053 UP000007798 UP000002282 UP000000304 UP000000803 UP000008711 UP000007801 UP000092461 UP000001292 UP000192221 UP000192223 UP000095301 UP000245037 UP000027135 UP000268350 UP000092553 UP000001070 UP000076858 UP000001819 UP000008744 UP000092462 UP000092443 UP000092460 UP000078200 UP000092444 UP000009192
UP000283053 UP000007798 UP000002282 UP000000304 UP000000803 UP000008711 UP000007801 UP000092461 UP000001292 UP000192221 UP000192223 UP000095301 UP000245037 UP000027135 UP000268350 UP000092553 UP000001070 UP000076858 UP000001819 UP000008744 UP000092462 UP000092443 UP000092460 UP000078200 UP000092444 UP000009192
Interpro
IPR040255
Non-specific_endonuclease
+ More
IPR001604 DNA/RNA_non-sp_Endonuclease
IPR020821 Extracellular_endonuc_su_A
IPR015324 Ribosomal_Rsm22-like
IPR029063 SAM-dependent_MTases
IPR038518 Glyco_hydro_63N_sf
IPR031631 Glyco_hydro_63N
IPR008928 6-hairpin_glycosidase_sf
IPR012341 6hp_glycosidase-like_sf
IPR031335 Glyco_hydro_63_C
IPR004888 Glycoside_hydrolase_63
IPR001604 DNA/RNA_non-sp_Endonuclease
IPR020821 Extracellular_endonuc_su_A
IPR015324 Ribosomal_Rsm22-like
IPR029063 SAM-dependent_MTases
IPR038518 Glyco_hydro_63N_sf
IPR031631 Glyco_hydro_63N
IPR008928 6-hairpin_glycosidase_sf
IPR012341 6hp_glycosidase-like_sf
IPR031335 Glyco_hydro_63_C
IPR004888 Glycoside_hydrolase_63
Gene 3D
ProteinModelPortal
A0A2H1V3T5
A0A1E1WMF9
A0A219LVV4
H9IV93
A0A2A4JNR7
A0A2W1BP50
+ More
Q08JX1 A0A2H1WP54 A0A2A4JML0 B7VCC3 A0A219LVV6 A0A2A4JP76 A0A194Q8Y6 A0A2W1BYI3 A0A1E1WTA0 A0A194RKW6 B7VCC2 F6K708 A0A2H1W493 A0A219LVU7 A0A219LVV5 A0A0N1IB53 A0A194Q8R2 A0A194Q8S9 A0A0N1IIN7 A0A212ETI7 A0A0N1ICG2 A0A212FE21 A0A2W1BV74 A0A0L7LLE8 A0A2A4JKE1 A0A219LWI3 H9JIU6 A0A0L7KR40 A0A2H1WT12 A0A0N1PH39 A0A3S2NR20 A0A219LVU2 A0A194Q7A2 A0A2A4K9C7 A0A0N0PDC8 A0A212FJT7 A0A2H1VP74 A0A2H1W9B5 B4N4S4 B4PH14 A0A0J9RZ63 B4QQB0 Q9VVV1 B3NDV9 B3M7Q7 A0A1B0CE89 A0A212EIS2 B4IFY0 T1P9F7 W8BUZ2 W8BEM3 A0A1W4UQR3 A0A1W4XR54 A0A1I8MCL2 A0A2P8ZMT4 A0A067RF87 A0A0P5K0P6 A0A0N8B5M4 A0A0P5W6R4 A0A0P5TF11 A0A0P6C109 A0A3B0JT56 A0A0M4EDJ3 B4IYD0 A0A1L8DXM9 A0A1L8DXI7 A0A1L8DPN4 A0A164USM9 Q29DJ9 B4H6U2 A0A1B0D5G0 A0A1A9YID0 A0A1B0C252 B4N4S6 A0A1A9UG98 D3TPG4 A0A0P5FLY5 A0A1B0FEL1 B4KZ79 Q29DK1 B4H6U4 A0A0P5L6U2 A0A0P5JV63 A0A0P5RYT1 A0A0P5S977 A0A0P5NE77 A0A0P5S216 A0A0N8CAS7 A0A0P5NWM0 A0A0P5NP42 A0A0P5S6M9
Q08JX1 A0A2H1WP54 A0A2A4JML0 B7VCC3 A0A219LVV6 A0A2A4JP76 A0A194Q8Y6 A0A2W1BYI3 A0A1E1WTA0 A0A194RKW6 B7VCC2 F6K708 A0A2H1W493 A0A219LVU7 A0A219LVV5 A0A0N1IB53 A0A194Q8R2 A0A194Q8S9 A0A0N1IIN7 A0A212ETI7 A0A0N1ICG2 A0A212FE21 A0A2W1BV74 A0A0L7LLE8 A0A2A4JKE1 A0A219LWI3 H9JIU6 A0A0L7KR40 A0A2H1WT12 A0A0N1PH39 A0A3S2NR20 A0A219LVU2 A0A194Q7A2 A0A2A4K9C7 A0A0N0PDC8 A0A212FJT7 A0A2H1VP74 A0A2H1W9B5 B4N4S4 B4PH14 A0A0J9RZ63 B4QQB0 Q9VVV1 B3NDV9 B3M7Q7 A0A1B0CE89 A0A212EIS2 B4IFY0 T1P9F7 W8BUZ2 W8BEM3 A0A1W4UQR3 A0A1W4XR54 A0A1I8MCL2 A0A2P8ZMT4 A0A067RF87 A0A0P5K0P6 A0A0N8B5M4 A0A0P5W6R4 A0A0P5TF11 A0A0P6C109 A0A3B0JT56 A0A0M4EDJ3 B4IYD0 A0A1L8DXM9 A0A1L8DXI7 A0A1L8DPN4 A0A164USM9 Q29DJ9 B4H6U2 A0A1B0D5G0 A0A1A9YID0 A0A1B0C252 B4N4S6 A0A1A9UG98 D3TPG4 A0A0P5FLY5 A0A1B0FEL1 B4KZ79 Q29DK1 B4H6U4 A0A0P5L6U2 A0A0P5JV63 A0A0P5RYT1 A0A0P5S977 A0A0P5NE77 A0A0P5S216 A0A0N8CAS7 A0A0P5NWM0 A0A0P5NP42 A0A0P5S6M9
Ontologies
GO
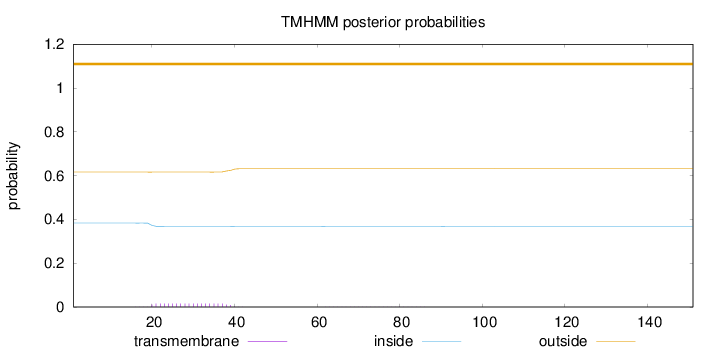
Topology
Length:
151
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.31144
Exp number, first 60 AAs:
0.30238
Total prob of N-in:
0.38359
outside
1 - 151
Population Genetic Test Statistics
Pi
191.478231
Theta
172.005148
Tajima's D
0.23215
CLR
0
CSRT
0.430428478576071
Interpretation
Uncertain
