Gene
KWMTBOMO07902
Pre Gene Modal
BGIBMGA000966
Annotation
PREDICTED:_leucine-rich_repeat-containing_protein_70_[Bombyx_mori]
Full name
Phosphorylase b kinase regulatory subunit
Location in the cell
PlasmaMembrane Reliability : 3.163
Sequence
CDS
ATGGCGCCCGCCTTTACATTCAGAGGGGTGGTGCGGTGGCTTGTGACGGGGATCGCTTGGGGGATGCTGTTTAGCTTGAGTCAAGCTCACACACCACCGTTCACGCCTTTGGCCACATTGACGCCGCCGTGTCCCACTCACTGCATTTGCCTGTCGCAATCTCAGGTGGTGTGTAATAGCGCTGCACTTCGCGGCGTTCCATCTGGCTTGAGCCCTTCTGTGACCCAGCTATCTCTATCGCGCGCAGACCTACGTGTTCTACGTTCTGATGCTTTTGCGCATCTTCGACAGTTGCGTCGTCTTGCTCTAGATGCCTGTAATCTAACACGTATTCGACCCTTTGCATTCCGAGGCCTGCCCAGACTTCGTGAGCTTTACATCCAGCACACGCCGTTGGCCACGGTCGATTCTTTCGCGTTTGCTGCTCTTCAAAATATAACTTCAATAGTTTTAACGCATAACAGGATAGCACAGATCGAAAGTTATGCATTTGCAGGAACAAACTTTATAAATCTAATTTCTTTACGAAACAATCCTATTAAACGCATTCTGGTGCATGCGTTCTCTGGACTCAACGACGTGGGGCAAATTGAGCTACCATCAGGGATAAGAACTATAGAACCACAAGCGTTCGCGGGCTTAGAGGGGATCGGCGTATTGGAGCTTGCGTTTATGGATTTGCCGTCATTACTCCCGGACACTTTCCACGGACTGATACGAGTCGGACGCCTTTCCCTACGCGAGTCTGATCTAGGTATCATTAAAGTTGGATCATTCGACGGACTACAGCACGTGGATACGCTTGAAATATGTAACAACAAAATCGATGGTATCGAAGAGCTGTCATTGGTCCAGAATAATAGTGTGCGAGTATTTAAATTAACCGGCAACCATATGTTAGAATCCCCGGAGGCTGTCGTGCTAGAAGTTGAGAATATTATTATCCGAGGGAATCATTTGCCGTGCGAGTGTGGACGTGATCCATTCACCAATCCTTTGGCGTTGAGTCAAGATTTTGCGGAAGAAAATTATTGTATATCACCGTTAAAAGTGCGTGGTCGAAGTCTTGCTAGTGCGACGAGTGGCAGCGCTGGTTGTCGTGGGGAGACTGGTGGCAGTGCACGGGCGCGAGCTTCTGCTGGTGCGGGTCCTGCCCGGAATGTCTACTCTCTAGCTGAATTAAACCTAGCTTCCATGGCTGCAGTCGTTGCATTCCTAGCGACTTGA
Protein
MAPAFTFRGVVRWLVTGIAWGMLFSLSQAHTPPFTPLATLTPPCPTHCICLSQSQVVCNSAALRGVPSGLSPSVTQLSLSRADLRVLRSDAFAHLRQLRRLALDACNLTRIRPFAFRGLPRLRELYIQHTPLATVDSFAFAALQNITSIVLTHNRIAQIESYAFAGTNFINLISLRNNPIKRILVHAFSGLNDVGQIELPSGIRTIEPQAFAGLEGIGVLELAFMDLPSLLPDTFHGLIRVGRLSLRESDLGIIKVGSFDGLQHVDTLEICNNKIDGIEELSLVQNNSVRVFKLTGNHMLESPEAVVLEVENIIIRGNHLPCECGRDPFTNPLALSQDFAEENYCISPLKVRGRSLASATSGSAGCRGETGGSARARASAGAGPARNVYSLAELNLASMAAVVAFLAT
Summary
Description
Phosphorylase b kinase catalyzes the phosphorylation of serine in certain substrates, including troponin I.
Similarity
Belongs to the phosphorylase b kinase regulatory chain family.
Uniprot
H9IUN6
A0A2W1BRU3
A0A2H1W6Z7
A0A2A4J2M3
A0A212FLN7
A0A3S2TLQ2
+ More
A0A194RLW1 A0A1B0CWP7 A0A0P4VGS3 T1I9G3 A0A182FQA4 A0A2R7X4Y5 A0A182RBU1 W5JJV3 A0A182LZV6 A0A182PIZ3 A0A084W949 A0A182N6X9 A0A182XX76 A0A182JIG8 A0A336MWD4 A0A182VQB2 A0A1A9WA83 A0A1A9XKF2 A0A1B0AUE1 A0A182QQ76 A0A1B0FKC5 A0A1A9VND2 A0A1B0AFF7 A0A182SEQ6 A0A1J1HMW1 Q7QK10 A0A1I8MVX8 A0A1I8NVK9 A0A0L0C7E7 Q29GW9 B3MQH4 B4NCN2 A0A3B0J9A2 E1NZD8 B4L7W2 B3NWH7 A0A1S4FR38 B4I659 B4M9Z5 A0A1W4V430 F5HIW3 Q9VR83 A0A182GBG2 A0A0C9Q1P7 A0A1I8MW91 A0A2J7Q6R9 B4JKB1 B0WF36 V9IJ23 A0A310S8D0 A0A182TUJ0 A0A1A9VND6 A0A2A3E3Q1 A0A1I8P7P3 A0A1B0AFF3 A0A1A9XKF5 A0A0M8ZY71 A0A0L0C4Y6 A0A1B0AUD5 A0A1A9WA82 Q16S33 A0A0M5J3F7 A0A2S2R101 F6JQ79 A0A151IKI4 F6JQ81 E2AEB5 F6JQ71 F6JQ72 F6JQ75 F6JQ80 A0A067RLB4 E9IW67 A0A232FB54 A0A158N9J9 J9KAC1 A0A0L7R0V5 A0A2H8U0K9 E0VBF8 A0A026WE81 A0A3L8E4Y5 N6T885 A0A1B0FNY5 A0A154PAL1 A0A2S2P621 A0A151JXI2 E2C8R5 A0A195E288 F4X7T3 B4GV34
A0A194RLW1 A0A1B0CWP7 A0A0P4VGS3 T1I9G3 A0A182FQA4 A0A2R7X4Y5 A0A182RBU1 W5JJV3 A0A182LZV6 A0A182PIZ3 A0A084W949 A0A182N6X9 A0A182XX76 A0A182JIG8 A0A336MWD4 A0A182VQB2 A0A1A9WA83 A0A1A9XKF2 A0A1B0AUE1 A0A182QQ76 A0A1B0FKC5 A0A1A9VND2 A0A1B0AFF7 A0A182SEQ6 A0A1J1HMW1 Q7QK10 A0A1I8MVX8 A0A1I8NVK9 A0A0L0C7E7 Q29GW9 B3MQH4 B4NCN2 A0A3B0J9A2 E1NZD8 B4L7W2 B3NWH7 A0A1S4FR38 B4I659 B4M9Z5 A0A1W4V430 F5HIW3 Q9VR83 A0A182GBG2 A0A0C9Q1P7 A0A1I8MW91 A0A2J7Q6R9 B4JKB1 B0WF36 V9IJ23 A0A310S8D0 A0A182TUJ0 A0A1A9VND6 A0A2A3E3Q1 A0A1I8P7P3 A0A1B0AFF3 A0A1A9XKF5 A0A0M8ZY71 A0A0L0C4Y6 A0A1B0AUD5 A0A1A9WA82 Q16S33 A0A0M5J3F7 A0A2S2R101 F6JQ79 A0A151IKI4 F6JQ81 E2AEB5 F6JQ71 F6JQ72 F6JQ75 F6JQ80 A0A067RLB4 E9IW67 A0A232FB54 A0A158N9J9 J9KAC1 A0A0L7R0V5 A0A2H8U0K9 E0VBF8 A0A026WE81 A0A3L8E4Y5 N6T885 A0A1B0FNY5 A0A154PAL1 A0A2S2P621 A0A151JXI2 E2C8R5 A0A195E288 F4X7T3 B4GV34
Pubmed
19121390
28756777
22118469
26354079
27129103
20920257
+ More
23761445 24438588 25244985 12364791 14747013 17210077 25315136 26108605 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 17510324 21383965 20798317 24845553 21282665 28648823 21347285 20566863 24508170 30249741 23537049 21719571
23761445 24438588 25244985 12364791 14747013 17210077 25315136 26108605 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 17510324 21383965 20798317 24845553 21282665 28648823 21347285 20566863 24508170 30249741 23537049 21719571
EMBL
BABH01000778
KZ149978
PZC75877.1
ODYU01006735
SOQ48850.1
NWSH01003553
+ More
PCG66119.1 AGBW02007761 OWR54610.1 RSAL01000063 RVE49518.1 KQ460205 KPJ16971.1 AJWK01032736 GDKW01003071 JAI53524.1 ACPB03002809 KK856846 PTY26480.1 ADMH02000923 ETN64657.1 AXCM01002961 ATLV01021605 KE525320 KFB46743.1 UFQS01002932 UFQT01001375 UFQT01002932 SSX14921.1 SSX30258.1 JXJN01003611 AXCN02000313 CCAG010003440 CVRI01000005 CRK87806.1 AAAB01008805 EAA03896.5 JRES01000909 KNC27349.1 CH379064 EAL31990.2 CH902621 EDV44600.2 CH964239 EDW82591.1 OUUW01000003 SPP78485.1 BT125687 ADO16253.1 CH933814 EDW05537.1 CH954180 EDV46797.2 CH480823 EDW56265.1 CH940655 EDW66054.2 EGK96224.1 AE014298 AAF50921.2 AGB95581.1 AGB95582.1 AGB95583.1 JXUM01052517 JXUM01052518 JXUM01052519 JXUM01102855 JXUM01102856 JXUM01102857 KQ564758 KQ561737 KXJ71777.1 KXJ77687.1 GBYB01007853 JAG77620.1 NEVH01017454 PNF24280.1 CH916370 EDW00014.1 DS231914 EDS25981.1 JR049881 AEY61108.1 KQ769812 OAD52828.1 KZ288439 PBC25721.1 KQ435826 KOX71999.1 KNC27345.1 JXJN01003608 CH477686 EAT37266.1 CP012528 ALC49386.1 GGMS01013849 MBY83052.1 GQ326592 ACU26566.1 KQ977203 KYN04965.1 GQ326594 ACU26568.1 GL438827 EFN68348.1 GQ326584 ACU26558.1 GQ326585 ACU26559.1 GQ326588 ACU26562.1 GQ326583 GQ326586 GQ326587 GQ326589 GQ326590 GQ326591 GQ326593 GQ326595 GQ326596 GQ326597 GQ326598 GQ326600 GQ326601 GQ326602 ACU26557.1 ACU26560.1 ACU26561.1 ACU26563.1 ACU26564.1 ACU26565.1 ACU26567.1 ACU26569.1 ACU26570.1 ACU26571.1 ACU26572.1 ACU26573.1 ACU26574.1 ACU26575.1 KK852444 KDR23818.1 GL766449 EFZ15179.1 NNAY01000554 OXU27699.1 ADTU01009673 ABLF02024987 ABLF02024988 KQ414670 KOC64426.1 GFXV01007904 MBW19709.1 DS235030 EEB10714.1 KK107260 EZA54233.1 QOIP01000001 RLU27008.1 APGK01040277 APGK01040278 KB740978 ENN76444.1 CCAG010013682 KQ434857 KZC08883.1 GGMR01012029 MBY24648.1 KQ981629 KYN38968.1 GL453693 EFN75661.1 KQ979814 KYN19032.1 GL888891 EGI57489.1 CH479192 EDW26571.1
PCG66119.1 AGBW02007761 OWR54610.1 RSAL01000063 RVE49518.1 KQ460205 KPJ16971.1 AJWK01032736 GDKW01003071 JAI53524.1 ACPB03002809 KK856846 PTY26480.1 ADMH02000923 ETN64657.1 AXCM01002961 ATLV01021605 KE525320 KFB46743.1 UFQS01002932 UFQT01001375 UFQT01002932 SSX14921.1 SSX30258.1 JXJN01003611 AXCN02000313 CCAG010003440 CVRI01000005 CRK87806.1 AAAB01008805 EAA03896.5 JRES01000909 KNC27349.1 CH379064 EAL31990.2 CH902621 EDV44600.2 CH964239 EDW82591.1 OUUW01000003 SPP78485.1 BT125687 ADO16253.1 CH933814 EDW05537.1 CH954180 EDV46797.2 CH480823 EDW56265.1 CH940655 EDW66054.2 EGK96224.1 AE014298 AAF50921.2 AGB95581.1 AGB95582.1 AGB95583.1 JXUM01052517 JXUM01052518 JXUM01052519 JXUM01102855 JXUM01102856 JXUM01102857 KQ564758 KQ561737 KXJ71777.1 KXJ77687.1 GBYB01007853 JAG77620.1 NEVH01017454 PNF24280.1 CH916370 EDW00014.1 DS231914 EDS25981.1 JR049881 AEY61108.1 KQ769812 OAD52828.1 KZ288439 PBC25721.1 KQ435826 KOX71999.1 KNC27345.1 JXJN01003608 CH477686 EAT37266.1 CP012528 ALC49386.1 GGMS01013849 MBY83052.1 GQ326592 ACU26566.1 KQ977203 KYN04965.1 GQ326594 ACU26568.1 GL438827 EFN68348.1 GQ326584 ACU26558.1 GQ326585 ACU26559.1 GQ326588 ACU26562.1 GQ326583 GQ326586 GQ326587 GQ326589 GQ326590 GQ326591 GQ326593 GQ326595 GQ326596 GQ326597 GQ326598 GQ326600 GQ326601 GQ326602 ACU26557.1 ACU26560.1 ACU26561.1 ACU26563.1 ACU26564.1 ACU26565.1 ACU26567.1 ACU26569.1 ACU26570.1 ACU26571.1 ACU26572.1 ACU26573.1 ACU26574.1 ACU26575.1 KK852444 KDR23818.1 GL766449 EFZ15179.1 NNAY01000554 OXU27699.1 ADTU01009673 ABLF02024987 ABLF02024988 KQ414670 KOC64426.1 GFXV01007904 MBW19709.1 DS235030 EEB10714.1 KK107260 EZA54233.1 QOIP01000001 RLU27008.1 APGK01040277 APGK01040278 KB740978 ENN76444.1 CCAG010013682 KQ434857 KZC08883.1 GGMR01012029 MBY24648.1 KQ981629 KYN38968.1 GL453693 EFN75661.1 KQ979814 KYN19032.1 GL888891 EGI57489.1 CH479192 EDW26571.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000092461
+ More
UP000015103 UP000069272 UP000075900 UP000000673 UP000075883 UP000075885 UP000030765 UP000075884 UP000076408 UP000075880 UP000075920 UP000091820 UP000092443 UP000092460 UP000075886 UP000092444 UP000078200 UP000092445 UP000075901 UP000183832 UP000007062 UP000095301 UP000095300 UP000037069 UP000001819 UP000007801 UP000007798 UP000268350 UP000009192 UP000008711 UP000001292 UP000008792 UP000192221 UP000000803 UP000069940 UP000249989 UP000235965 UP000001070 UP000002320 UP000075902 UP000242457 UP000053105 UP000008820 UP000092553 UP000078542 UP000000311 UP000027135 UP000215335 UP000005205 UP000007819 UP000053825 UP000009046 UP000053097 UP000279307 UP000019118 UP000076502 UP000078541 UP000008237 UP000078492 UP000007755 UP000008744
UP000015103 UP000069272 UP000075900 UP000000673 UP000075883 UP000075885 UP000030765 UP000075884 UP000076408 UP000075880 UP000075920 UP000091820 UP000092443 UP000092460 UP000075886 UP000092444 UP000078200 UP000092445 UP000075901 UP000183832 UP000007062 UP000095301 UP000095300 UP000037069 UP000001819 UP000007801 UP000007798 UP000268350 UP000009192 UP000008711 UP000001292 UP000008792 UP000192221 UP000000803 UP000069940 UP000249989 UP000235965 UP000001070 UP000002320 UP000075902 UP000242457 UP000053105 UP000008820 UP000092553 UP000078542 UP000000311 UP000027135 UP000215335 UP000005205 UP000007819 UP000053825 UP000009046 UP000053097 UP000279307 UP000019118 UP000076502 UP000078541 UP000008237 UP000078492 UP000007755 UP000008744
Interpro
SUPFAM
SSF48208
SSF48208
Gene 3D
ProteinModelPortal
H9IUN6
A0A2W1BRU3
A0A2H1W6Z7
A0A2A4J2M3
A0A212FLN7
A0A3S2TLQ2
+ More
A0A194RLW1 A0A1B0CWP7 A0A0P4VGS3 T1I9G3 A0A182FQA4 A0A2R7X4Y5 A0A182RBU1 W5JJV3 A0A182LZV6 A0A182PIZ3 A0A084W949 A0A182N6X9 A0A182XX76 A0A182JIG8 A0A336MWD4 A0A182VQB2 A0A1A9WA83 A0A1A9XKF2 A0A1B0AUE1 A0A182QQ76 A0A1B0FKC5 A0A1A9VND2 A0A1B0AFF7 A0A182SEQ6 A0A1J1HMW1 Q7QK10 A0A1I8MVX8 A0A1I8NVK9 A0A0L0C7E7 Q29GW9 B3MQH4 B4NCN2 A0A3B0J9A2 E1NZD8 B4L7W2 B3NWH7 A0A1S4FR38 B4I659 B4M9Z5 A0A1W4V430 F5HIW3 Q9VR83 A0A182GBG2 A0A0C9Q1P7 A0A1I8MW91 A0A2J7Q6R9 B4JKB1 B0WF36 V9IJ23 A0A310S8D0 A0A182TUJ0 A0A1A9VND6 A0A2A3E3Q1 A0A1I8P7P3 A0A1B0AFF3 A0A1A9XKF5 A0A0M8ZY71 A0A0L0C4Y6 A0A1B0AUD5 A0A1A9WA82 Q16S33 A0A0M5J3F7 A0A2S2R101 F6JQ79 A0A151IKI4 F6JQ81 E2AEB5 F6JQ71 F6JQ72 F6JQ75 F6JQ80 A0A067RLB4 E9IW67 A0A232FB54 A0A158N9J9 J9KAC1 A0A0L7R0V5 A0A2H8U0K9 E0VBF8 A0A026WE81 A0A3L8E4Y5 N6T885 A0A1B0FNY5 A0A154PAL1 A0A2S2P621 A0A151JXI2 E2C8R5 A0A195E288 F4X7T3 B4GV34
A0A194RLW1 A0A1B0CWP7 A0A0P4VGS3 T1I9G3 A0A182FQA4 A0A2R7X4Y5 A0A182RBU1 W5JJV3 A0A182LZV6 A0A182PIZ3 A0A084W949 A0A182N6X9 A0A182XX76 A0A182JIG8 A0A336MWD4 A0A182VQB2 A0A1A9WA83 A0A1A9XKF2 A0A1B0AUE1 A0A182QQ76 A0A1B0FKC5 A0A1A9VND2 A0A1B0AFF7 A0A182SEQ6 A0A1J1HMW1 Q7QK10 A0A1I8MVX8 A0A1I8NVK9 A0A0L0C7E7 Q29GW9 B3MQH4 B4NCN2 A0A3B0J9A2 E1NZD8 B4L7W2 B3NWH7 A0A1S4FR38 B4I659 B4M9Z5 A0A1W4V430 F5HIW3 Q9VR83 A0A182GBG2 A0A0C9Q1P7 A0A1I8MW91 A0A2J7Q6R9 B4JKB1 B0WF36 V9IJ23 A0A310S8D0 A0A182TUJ0 A0A1A9VND6 A0A2A3E3Q1 A0A1I8P7P3 A0A1B0AFF3 A0A1A9XKF5 A0A0M8ZY71 A0A0L0C4Y6 A0A1B0AUD5 A0A1A9WA82 Q16S33 A0A0M5J3F7 A0A2S2R101 F6JQ79 A0A151IKI4 F6JQ81 E2AEB5 F6JQ71 F6JQ72 F6JQ75 F6JQ80 A0A067RLB4 E9IW67 A0A232FB54 A0A158N9J9 J9KAC1 A0A0L7R0V5 A0A2H8U0K9 E0VBF8 A0A026WE81 A0A3L8E4Y5 N6T885 A0A1B0FNY5 A0A154PAL1 A0A2S2P621 A0A151JXI2 E2C8R5 A0A195E288 F4X7T3 B4GV34
PDB
3ZYJ
E-value=4.4781e-18,
Score=224
Ontologies
GO
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 29,
Likelihood: 0.913880
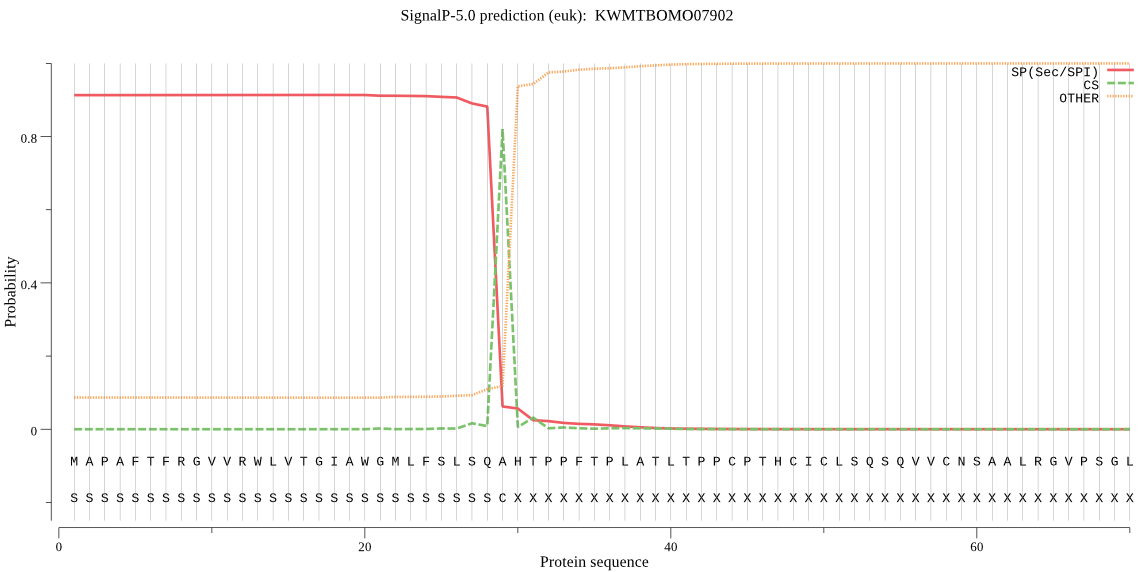
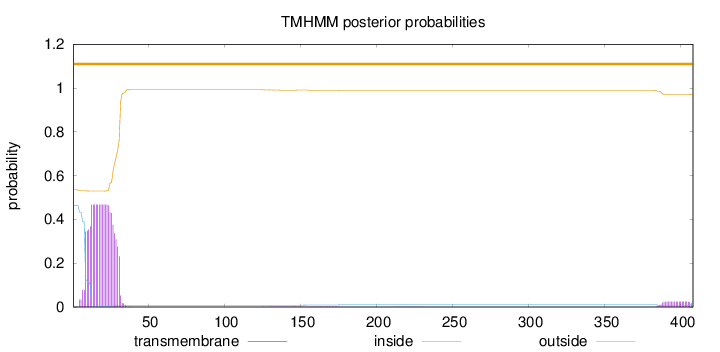
Length:
408
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.31799
Exp number, first 60 AAs:
9.73935
Total prob of N-in:
0.46506
outside
1 - 408
Population Genetic Test Statistics
Pi
255.24537
Theta
185.175237
Tajima's D
0.656773
CLR
0.188179
CSRT
0.562821858907055
Interpretation
Uncertain
