Gene
KWMTBOMO07897 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001185
Annotation
N-cetylglucosamine-6-phosphate_deacetylase_[Spodoptera_litura]
Full name
N-acetylglucosamine-6-phosphate deacetylase
Location in the cell
Cytoplasmic Reliability : 1.584 Extracellular Reliability : 1.635
Sequence
CDS
ATGTATAGCGAGCGGGCATACGGCCGCGAGAGTGACGTACCGTTCCATCATCGTGATCCAGGATTAGTAGGGTTACTAGCTTCTACGATGAAAAAACAAGTATATTATGGTATAATAGCTGATGGTATCCACACTCATCCAGCAGCGCTCAGAATAGCAAACAGAACTAATCCCGAAGGTTTAGTACTAGTAAGTGATGCAGTGCCAGCGCAGGGACTACCGGATGGTAACCATCGCATTGGACCGCAAATCGTGACGGTAGAAGACGGTTGCGCTTACGTCACTGGAACGAAAACCCTCTGTGGAAGCACTACTGCCCTGGACTCCTGTATTGTGAAATTTAAAGACTCGATTGAATGTAGCTTGGAATATGCATTGGAAGCAGCCTCATTACATCCGGCTAAGGCTTTAGGAATTGATGACAAGAAAGGGAATTTAAACTTTGACAGTGACGCTGACTTCGTAATACTTCACCCTAAAACATTGAAAGTTCAATCCACCTGGATAGCTGGAGAATGTGTATATAAATGCCAGAACTAA
Protein
MYSERAYGRESDVPFHHRDPGLVGLLASTMKKQVYYGIIADGIHTHPAALRIANRTNPEGLVLVSDAVPAQGLPDGNHRIGPQIVTVEDGCAYVTGTKTLCGSTTALDSCIVKFKDSIECSLEYALEAASLHPAKALGIDDKKGNLNFDSDADFVILHPKTLKVQSTWIAGECVYKCQN
Summary
Description
Hydrolyzes the N-glycolyl group from N-glycolylglucosamine 6-phosphate (GlcNGc-6-P) in the N-glycolylneuraminic acid (Neu5Gc) degradation pathway.
Catalytic Activity
H2O + N-acetyl-D-glucosamine 6-phosphate = acetate + D-glucosamine 6-phosphate
Cofactor
a divalent metal cation
Similarity
Belongs to the metallo-dependent hydrolases superfamily. NagA family.
Belongs to the universal ribosomal protein uS11 family.
Belongs to the universal ribosomal protein uS11 family.
Keywords
Carbohydrate metabolism
Complete proteome
Hydrolase
Metal-binding
Reference proteome
Feature
chain N-acetylglucosamine-6-phosphate deacetylase
Uniprot
H9IVA5
A0A2W1BTB6
A0A2A4J876
S5GAZ8
A0A212FLN5
S4NND0
+ More
I4DRC2 I4DJ61 A0A194RI15 A0A2H1W6Y2 A0A194QAC2 A0A0L7L1U4 A0A2D4GE62 A0A182VGP9 A0A1B6JGT0 A0A182SHB0 A0A2D4Q028 Q7QBX3 A0A182PA19 A0A182FR77 A0A182XID6 A0A2M4AMN1 W5JF47 A0A1L8E1N9 A0A2D4GEC7 A0A182GIW6 A0A182K3S8 B4GV30 A0A1B6M6N7 A0A1S4F1X7 Q17I97 A0A182U7I7 A0A1Q3FGT0 A0A182JLI6 W8C6X2 A0A0A1WZU0 A0A0K8WLL3 A0A2D4IPM1 A0A1B0CB00 A0A1B6G3U7 Q29GW7 A0A1J1IRB1 A0A0K8RSL3 A0A2J7PFL1 E2AFM4 A0A158NY12 A0A1B0D4T6 A0A195B8J4 G3I2T8 A0A151ICY0 A0A195FW46 F4WNM6 A0A034VT64 A0A182HTC0 A0A2C9K4U3 A0A195ECC4 A0A151XBY1 A0A2C9K4S2 A0A182M5S2 B0WS55 A0A1B6CLI3 A0A0C9R8Z0 A0A0L7R617 A0A3B0JXJ4 A0A1W4VF90 A0A182VQ89 A0A2J7PFM1 B3NWH2 A0A0K8WIC2 B4NCN7 J9JKI9 A0A139WIQ9 A0A0Q9XFY2 A0A182Y7A4 A0A0Q9XQE4 A0A3Q3N7S5 A0A182RCV7 A0A2J7PFK9 A0A061I330 U3JQK5 X2JFY0 Q9VR81 A0A1B6JIX3 A0A0B4J2M4 A0A336LUT2 A0A091UIU3 A0A2Y9DF13 E1JJR9 B4PY15 A0A226MVY0 A0A1I8NLQ4 A0A0P4WHL1 G3TV02 B4L7W7 A0A1A6GDM9 G3TG18 K1QXA4 A0A154PQZ8 A0A091WH03 I3MKZ4
I4DRC2 I4DJ61 A0A194RI15 A0A2H1W6Y2 A0A194QAC2 A0A0L7L1U4 A0A2D4GE62 A0A182VGP9 A0A1B6JGT0 A0A182SHB0 A0A2D4Q028 Q7QBX3 A0A182PA19 A0A182FR77 A0A182XID6 A0A2M4AMN1 W5JF47 A0A1L8E1N9 A0A2D4GEC7 A0A182GIW6 A0A182K3S8 B4GV30 A0A1B6M6N7 A0A1S4F1X7 Q17I97 A0A182U7I7 A0A1Q3FGT0 A0A182JLI6 W8C6X2 A0A0A1WZU0 A0A0K8WLL3 A0A2D4IPM1 A0A1B0CB00 A0A1B6G3U7 Q29GW7 A0A1J1IRB1 A0A0K8RSL3 A0A2J7PFL1 E2AFM4 A0A158NY12 A0A1B0D4T6 A0A195B8J4 G3I2T8 A0A151ICY0 A0A195FW46 F4WNM6 A0A034VT64 A0A182HTC0 A0A2C9K4U3 A0A195ECC4 A0A151XBY1 A0A2C9K4S2 A0A182M5S2 B0WS55 A0A1B6CLI3 A0A0C9R8Z0 A0A0L7R617 A0A3B0JXJ4 A0A1W4VF90 A0A182VQ89 A0A2J7PFM1 B3NWH2 A0A0K8WIC2 B4NCN7 J9JKI9 A0A139WIQ9 A0A0Q9XFY2 A0A182Y7A4 A0A0Q9XQE4 A0A3Q3N7S5 A0A182RCV7 A0A2J7PFK9 A0A061I330 U3JQK5 X2JFY0 Q9VR81 A0A1B6JIX3 A0A0B4J2M4 A0A336LUT2 A0A091UIU3 A0A2Y9DF13 E1JJR9 B4PY15 A0A226MVY0 A0A1I8NLQ4 A0A0P4WHL1 G3TV02 B4L7W7 A0A1A6GDM9 G3TG18 K1QXA4 A0A154PQZ8 A0A091WH03 I3MKZ4
EC Number
3.5.1.25
Pubmed
19121390
28756777
22118469
23622113
22651552
26354079
+ More
26227816 12364791 14747013 17210077 20920257 23761445 26483478 17994087 17510324 24495485 25830018 15632085 25727380 20798317 21347285 21804562 21719571 25348373 15562597 18362917 19820115 25244985 23929341 29704459 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 17550304 18057021 22992520
26227816 12364791 14747013 17210077 20920257 23761445 26483478 17994087 17510324 24495485 25830018 15632085 25727380 20798317 21347285 21804562 21719571 25348373 15562597 18362917 19820115 25244985 23929341 29704459 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 17550304 18057021 22992520
EMBL
BABH01000784
BABH01000785
KZ149978
PZC75876.1
NWSH01002427
PCG68327.1
+ More
KC832291 AGQ53951.1 AGBW02007761 OWR54609.1 GAIX01012304 JAA80256.1 AK405059 BAM20462.1 AK401329 BAM17951.1 KQ460205 KPJ16970.1 ODYU01006735 SOQ48849.1 KQ459299 KPJ01945.1 JTDY01003645 KOB69229.1 IACJ01123622 LAA58061.1 GECU01013216 GECU01009322 JAS94490.1 JAS98384.1 IACN01100870 LAB63567.1 AAAB01008859 EAA08223.5 GGFK01008734 MBW42055.1 ADMH02001587 ETN61953.1 GFDF01001456 JAV12628.1 IACJ01123621 LAA58058.1 JXUM01012630 JXUM01012631 JXUM01012632 KQ560358 KXJ82845.1 CH479192 EDW26567.1 GEBQ01008387 JAT31590.1 CH477241 EAT46400.1 GFDL01008284 JAV26761.1 AXCP01008967 GAMC01000877 JAC05679.1 GBXI01009950 JAD04342.1 GDHF01000589 JAI51725.1 IACK01119929 LAA86158.1 AJWK01004691 GECZ01012654 JAS57115.1 CH379064 EAL31992.2 CVRI01000058 CRL02688.1 GBKC01002274 JAG43796.1 NEVH01025655 PNF15119.1 GL439118 EFN67775.1 ADTU01002870 AJVK01024863 KQ976565 KYM80514.1 JH001143 EGW09759.1 KQ978048 KYM97798.1 KQ981215 KYN44542.1 GL888237 EGI64219.1 GAKP01012431 JAC46521.1 APCN01000438 KQ979074 KYN22761.1 KQ982316 KYQ57820.1 AXCM01001115 DS232064 EDS33674.1 GEDC01023157 JAS14141.1 GBYB01003256 JAG73023.1 KQ414648 KOC66315.1 OUUW01000003 SPP78489.1 PNF15118.1 CH954180 EDV46792.1 GDHF01001520 JAI50794.1 CH964239 EDW82596.1 ABLF02029453 KQ971338 KYB27794.1 CH933814 KRG07430.1 KRG07428.1 PNF15120.1 KE682131 RAZU01000243 ERE67775.1 RLQ62918.1 AGTO01020847 AE014298 AHN59958.1 AY089672 GECU01008460 JAS99246.1 UFQT01000205 UFQT01001174 SSX21694.1 SSX29306.1 KL409574 KFQ90631.1 ACZ95338.1 CM000162 EDX02987.1 MCFN01000402 OXB59280.1 GDIP01255160 LRGB01001581 JAI68241.1 KZS11258.1 EDW05542.1 LZPO01097222 OBS64356.1 JH818609 EKC35804.1 KQ435066 KZC14336.1 KK735490 KFR14400.1 AGTP01095340
KC832291 AGQ53951.1 AGBW02007761 OWR54609.1 GAIX01012304 JAA80256.1 AK405059 BAM20462.1 AK401329 BAM17951.1 KQ460205 KPJ16970.1 ODYU01006735 SOQ48849.1 KQ459299 KPJ01945.1 JTDY01003645 KOB69229.1 IACJ01123622 LAA58061.1 GECU01013216 GECU01009322 JAS94490.1 JAS98384.1 IACN01100870 LAB63567.1 AAAB01008859 EAA08223.5 GGFK01008734 MBW42055.1 ADMH02001587 ETN61953.1 GFDF01001456 JAV12628.1 IACJ01123621 LAA58058.1 JXUM01012630 JXUM01012631 JXUM01012632 KQ560358 KXJ82845.1 CH479192 EDW26567.1 GEBQ01008387 JAT31590.1 CH477241 EAT46400.1 GFDL01008284 JAV26761.1 AXCP01008967 GAMC01000877 JAC05679.1 GBXI01009950 JAD04342.1 GDHF01000589 JAI51725.1 IACK01119929 LAA86158.1 AJWK01004691 GECZ01012654 JAS57115.1 CH379064 EAL31992.2 CVRI01000058 CRL02688.1 GBKC01002274 JAG43796.1 NEVH01025655 PNF15119.1 GL439118 EFN67775.1 ADTU01002870 AJVK01024863 KQ976565 KYM80514.1 JH001143 EGW09759.1 KQ978048 KYM97798.1 KQ981215 KYN44542.1 GL888237 EGI64219.1 GAKP01012431 JAC46521.1 APCN01000438 KQ979074 KYN22761.1 KQ982316 KYQ57820.1 AXCM01001115 DS232064 EDS33674.1 GEDC01023157 JAS14141.1 GBYB01003256 JAG73023.1 KQ414648 KOC66315.1 OUUW01000003 SPP78489.1 PNF15118.1 CH954180 EDV46792.1 GDHF01001520 JAI50794.1 CH964239 EDW82596.1 ABLF02029453 KQ971338 KYB27794.1 CH933814 KRG07430.1 KRG07428.1 PNF15120.1 KE682131 RAZU01000243 ERE67775.1 RLQ62918.1 AGTO01020847 AE014298 AHN59958.1 AY089672 GECU01008460 JAS99246.1 UFQT01000205 UFQT01001174 SSX21694.1 SSX29306.1 KL409574 KFQ90631.1 ACZ95338.1 CM000162 EDX02987.1 MCFN01000402 OXB59280.1 GDIP01255160 LRGB01001581 JAI68241.1 KZS11258.1 EDW05542.1 LZPO01097222 OBS64356.1 JH818609 EKC35804.1 KQ435066 KZC14336.1 KK735490 KFR14400.1 AGTP01095340
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000075903 UP000075901 UP000007062 UP000075885 UP000069272 UP000076407 UP000000673 UP000069940 UP000249989 UP000075881 UP000008744 UP000008820 UP000075902 UP000075880 UP000092461 UP000001819 UP000183832 UP000235965 UP000000311 UP000005205 UP000092462 UP000078540 UP000001075 UP000078542 UP000078541 UP000007755 UP000075840 UP000076420 UP000078492 UP000075809 UP000075883 UP000002320 UP000053825 UP000268350 UP000192221 UP000075920 UP000008711 UP000007798 UP000007819 UP000007266 UP000009192 UP000076408 UP000261640 UP000075900 UP000030759 UP000273346 UP000016665 UP000000803 UP000005203 UP000053283 UP000248480 UP000002282 UP000198323 UP000095300 UP000076858 UP000007646 UP000092124 UP000005408 UP000076502 UP000053605 UP000005215
UP000075903 UP000075901 UP000007062 UP000075885 UP000069272 UP000076407 UP000000673 UP000069940 UP000249989 UP000075881 UP000008744 UP000008820 UP000075902 UP000075880 UP000092461 UP000001819 UP000183832 UP000235965 UP000000311 UP000005205 UP000092462 UP000078540 UP000001075 UP000078542 UP000078541 UP000007755 UP000075840 UP000076420 UP000078492 UP000075809 UP000075883 UP000002320 UP000053825 UP000268350 UP000192221 UP000075920 UP000008711 UP000007798 UP000007819 UP000007266 UP000009192 UP000076408 UP000261640 UP000075900 UP000030759 UP000273346 UP000016665 UP000000803 UP000005203 UP000053283 UP000248480 UP000002282 UP000198323 UP000095300 UP000076858 UP000007646 UP000092124 UP000005408 UP000076502 UP000053605 UP000005215
Pfam
Interpro
IPR011059
Metal-dep_hydrolase_composite
+ More
IPR006680 Amidohydro-rel
IPR003764 GlcNAc_6-P_deAcase
IPR032466 Metal_Hydrolase
IPR020568 Ribosomal_S5_D2-typ_fold
IPR033100 Rrp45
IPR036345 ExoRNase_PH_dom2_sf
IPR015847 ExoRNase_PH_dom2
IPR001247 ExoRNase_PH_dom1
IPR027408 PNPase/RNase_PH_dom_sf
IPR001971 Ribosomal_S11
IPR036967 Ribosomal_S11_sf
IPR018102 Ribosomal_S11_CS
IPR007588 Znf_FLYWCH
IPR006680 Amidohydro-rel
IPR003764 GlcNAc_6-P_deAcase
IPR032466 Metal_Hydrolase
IPR020568 Ribosomal_S5_D2-typ_fold
IPR033100 Rrp45
IPR036345 ExoRNase_PH_dom2_sf
IPR015847 ExoRNase_PH_dom2
IPR001247 ExoRNase_PH_dom1
IPR027408 PNPase/RNase_PH_dom_sf
IPR001971 Ribosomal_S11
IPR036967 Ribosomal_S11_sf
IPR018102 Ribosomal_S11_CS
IPR007588 Znf_FLYWCH
Gene 3D
ProteinModelPortal
H9IVA5
A0A2W1BTB6
A0A2A4J876
S5GAZ8
A0A212FLN5
S4NND0
+ More
I4DRC2 I4DJ61 A0A194RI15 A0A2H1W6Y2 A0A194QAC2 A0A0L7L1U4 A0A2D4GE62 A0A182VGP9 A0A1B6JGT0 A0A182SHB0 A0A2D4Q028 Q7QBX3 A0A182PA19 A0A182FR77 A0A182XID6 A0A2M4AMN1 W5JF47 A0A1L8E1N9 A0A2D4GEC7 A0A182GIW6 A0A182K3S8 B4GV30 A0A1B6M6N7 A0A1S4F1X7 Q17I97 A0A182U7I7 A0A1Q3FGT0 A0A182JLI6 W8C6X2 A0A0A1WZU0 A0A0K8WLL3 A0A2D4IPM1 A0A1B0CB00 A0A1B6G3U7 Q29GW7 A0A1J1IRB1 A0A0K8RSL3 A0A2J7PFL1 E2AFM4 A0A158NY12 A0A1B0D4T6 A0A195B8J4 G3I2T8 A0A151ICY0 A0A195FW46 F4WNM6 A0A034VT64 A0A182HTC0 A0A2C9K4U3 A0A195ECC4 A0A151XBY1 A0A2C9K4S2 A0A182M5S2 B0WS55 A0A1B6CLI3 A0A0C9R8Z0 A0A0L7R617 A0A3B0JXJ4 A0A1W4VF90 A0A182VQ89 A0A2J7PFM1 B3NWH2 A0A0K8WIC2 B4NCN7 J9JKI9 A0A139WIQ9 A0A0Q9XFY2 A0A182Y7A4 A0A0Q9XQE4 A0A3Q3N7S5 A0A182RCV7 A0A2J7PFK9 A0A061I330 U3JQK5 X2JFY0 Q9VR81 A0A1B6JIX3 A0A0B4J2M4 A0A336LUT2 A0A091UIU3 A0A2Y9DF13 E1JJR9 B4PY15 A0A226MVY0 A0A1I8NLQ4 A0A0P4WHL1 G3TV02 B4L7W7 A0A1A6GDM9 G3TG18 K1QXA4 A0A154PQZ8 A0A091WH03 I3MKZ4
I4DRC2 I4DJ61 A0A194RI15 A0A2H1W6Y2 A0A194QAC2 A0A0L7L1U4 A0A2D4GE62 A0A182VGP9 A0A1B6JGT0 A0A182SHB0 A0A2D4Q028 Q7QBX3 A0A182PA19 A0A182FR77 A0A182XID6 A0A2M4AMN1 W5JF47 A0A1L8E1N9 A0A2D4GEC7 A0A182GIW6 A0A182K3S8 B4GV30 A0A1B6M6N7 A0A1S4F1X7 Q17I97 A0A182U7I7 A0A1Q3FGT0 A0A182JLI6 W8C6X2 A0A0A1WZU0 A0A0K8WLL3 A0A2D4IPM1 A0A1B0CB00 A0A1B6G3U7 Q29GW7 A0A1J1IRB1 A0A0K8RSL3 A0A2J7PFL1 E2AFM4 A0A158NY12 A0A1B0D4T6 A0A195B8J4 G3I2T8 A0A151ICY0 A0A195FW46 F4WNM6 A0A034VT64 A0A182HTC0 A0A2C9K4U3 A0A195ECC4 A0A151XBY1 A0A2C9K4S2 A0A182M5S2 B0WS55 A0A1B6CLI3 A0A0C9R8Z0 A0A0L7R617 A0A3B0JXJ4 A0A1W4VF90 A0A182VQ89 A0A2J7PFM1 B3NWH2 A0A0K8WIC2 B4NCN7 J9JKI9 A0A139WIQ9 A0A0Q9XFY2 A0A182Y7A4 A0A0Q9XQE4 A0A3Q3N7S5 A0A182RCV7 A0A2J7PFK9 A0A061I330 U3JQK5 X2JFY0 Q9VR81 A0A1B6JIX3 A0A0B4J2M4 A0A336LUT2 A0A091UIU3 A0A2Y9DF13 E1JJR9 B4PY15 A0A226MVY0 A0A1I8NLQ4 A0A0P4WHL1 G3TV02 B4L7W7 A0A1A6GDM9 G3TG18 K1QXA4 A0A154PQZ8 A0A091WH03 I3MKZ4
PDB
2VHL
E-value=1.08487e-16,
Score=207
Ontologies
GO
Topology
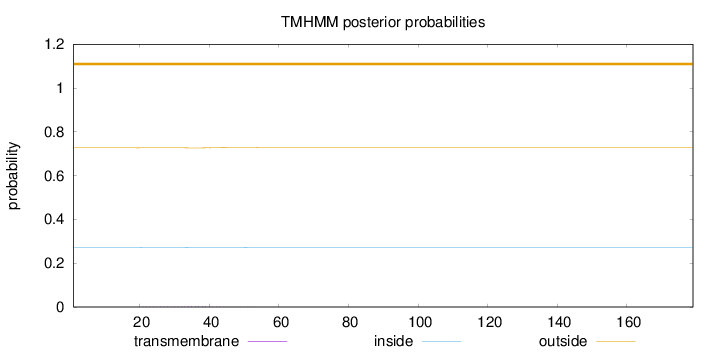
Length:
179
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05367
Exp number, first 60 AAs:
0.05323
Total prob of N-in:
0.27298
outside
1 - 179
Population Genetic Test Statistics
Pi
58.111085
Theta
64.253311
Tajima's D
0.092858
CLR
0.634029
CSRT
0.391980400979951
Interpretation
Uncertain
