Pre Gene Modal
BGIBMGA001188
Annotation
PREDICTED:_exosome_complex_component_RRP43-like_[Amyelois_transitella]
Full name
Exosome complex component RRP43
Alternative Name
Exosome component 8
Opa-interacting protein 2
Ribosomal RNA-processing protein 43
p9
Opa-interacting protein 2
Ribosomal RNA-processing protein 43
p9
Location in the cell
Cytoplasmic Reliability : 2.284
Sequence
CDS
ATGTCCGAAGTATATAAACTAATACATCCAGTAAAACATTTCAACGATTATATTTCTATCAAAACTAGACTAGACGGAAGAAACTTCGATGAACATCGGAACATAAAGTTAAATGTTAATTCCATAAAAACTGCAGATGCATCGGCCGTAGTTAAATGTGGTAATACAACAGTTGTATGTGGAATAAAACTGGAATTAGCAGTACCAAAAGCTGAAGAACCTGACTTAGGTTTTTTAATAACAAATGTGGAACTACCTGCTTTATGTTCTTCTAAGTATAGACCTGGCCCTCCTTCAGATTATGCACAAGTTATAAATAATATTGTATCAGAAATTATCAAAAATTCTAAATGCATTGATCTAAAAGATCTTTGCATTGTACCAGATAAATTAGCATGGGTTGTGTACTGTGATATGGTGTGTATTGACCATGACGGGAGCCTGGTTGATGCATGCATCATAGCACTGATAGCCAGCTTCAAAACATTACTATTACCATCTGTTAATTATGACACAGAAACAGAAGAAATAAAAGTAGAAACAAATGAGAGGATAAATCTCAAAATCCATGGACTACCGGTAGCTACAAGCTTTGCTGTGTACGAACATTTTCAAAGCCAAATAATATTATTAACTGATCCAACAAATTATGAAGAAGAGATGTGTGGAGGCAATGGATCAAATTTAATAATCTCCTACAACAAAGGCTTATTATGTGGCATTCAGAAATTGGGAGGTGGTAGTTTATCAACAGAAAATAATGAGGAAGCAATAAGATTAGCTGAACAACACACAATAGCAGTGGAGAAAGTAATCAATGCATGTTTAGAAGGAAAAGGAACAAATAAATAA
Protein
MSEVYKLIHPVKHFNDYISIKTRLDGRNFDEHRNIKLNVNSIKTADASAVVKCGNTTVVCGIKLELAVPKAEEPDLGFLITNVELPALCSSKYRPGPPSDYAQVINNIVSEIIKNSKCIDLKDLCIVPDKLAWVVYCDMVCIDHDGSLVDACIIALIASFKTLLLPSVNYDTETEEIKVETNERINLKIHGLPVATSFAVYEHFQSQIILLTDPTNYEEEMCGGNGSNLIISYNKGLLCGIQKLGGGSLSTENNEEAIRLAEQHTIAVEKVINACLEGKGTNK
Summary
Description
Non-catalytic component of the RNA exosome complex which has 3'->5' exoribonuclease activity and participates in a multitude of cellular RNA processing and degradation events. In the nucleus, the RNA exosome complex is involved in proper maturation of stable RNA species such as rRNA, snRNA and snoRNA, in the elimination of RNA processing by-products and non-coding 'pervasive' transcripts, such as antisense RNA species and promoter-upstream transcripts (PROMPTs), and of mRNAs with processing defects, thereby limiting or excluding their export to the cytoplasm. The RNA exosome may be involved in Ig class switch recombination (CSR) and/or Ig variable region somatic hypermutation (SHM) by targeting AICDA deamination activity to transcribed dsDNA substrates. In the cytoplasm, the RNA exosome complex is involved in general mRNA turnover and specifically degrades inherently unstable mRNAs containing AU-rich elements (AREs) within their 3' untranslated regions, and in RNA surveillance pathways, preventing translation of aberrant mRNAs. It seems to be involved in degradation of histone mRNA. The catalytic inactive RNA exosome core complex of 9 subunits (Exo-9) is proposed to play a pivotal role in the binding and presentation of RNA for ribonucleolysis, and to serve as a scaffold for the association with catalytic subunits and accessory proteins or complexes. EXOSC8 binds to ARE-containing RNAs.
Subunit
Component of the RNA exosome complex. Specifically part of the catalytically inactive RNA exosome core (Exo-9) complex which is believed to associate with catalytic subunits EXOSC10, and DIS3 or DIS3L in cytoplasmic- and nuclear-specific RNA exosome complex forms. Exo-9 is formed by a hexameric ring of RNase PH domain-containing subunits specifically containing the heterodimers EXOSC4-EXOSC9, EXOSC5-EXOSC8 and EXOSC6-EXOSC7, and peripheral S1 domain-containing components EXOSC1, EXOSC2 and EXOSC3 located on the top of the ring structure. Binds outer membrane protein opap from Neisseria gonorrhoeae.
Similarity
Belongs to the RNase PH family.
Keywords
3D-structure
Acetylation
Complete proteome
Cytoplasm
Disease mutation
Exosome
Neurodegeneration
Nucleus
Reference proteome
RNA-binding
rRNA processing
Feature
chain Exosome complex component RRP43
sequence variant In PCH1C; dbSNP:rs606231285.
sequence variant In PCH1C; dbSNP:rs606231285.
Uniprot
H9IVA8
A0A2A4JXZ4
A0A3S2NV38
S4P3V4
A0A0N1I9Y5
A0A2H1X1V6
+ More
A0A0L7LDK9 A0A194QAD1 A0A212FHC8 I4DPA4 A0A2J7PNN7 A0A067RJ27 A0A194RGK8 A0A2P8ZAR4 D2A0W4 A0A1B6CB03 A0A1B6L851 K7J7Z8 A0A1L8DXY1 A0A158NWG7 A0A1B6FP22 A0A151I515 A0A1L8DXS8 A0A232FEW2 A0A2A3E7S5 A0A151WYP5 E2BFG5 A0A088AJ30 F4WQ24 U4U0H3 A0A026W4F8 A0A1Y1NDX8 E2A2H2 A0A182P5V7 S4RI20 A0A1Y1NDI9 A0A182JYU8 A0A0L8I2M4 K4G0P6 Q17NN4 A0A195ECK5 A0A154PGF9 A0A0L7RBM2 A0A182HBH1 D1MLN6 A0A182WZT4 Q7Q0D9 A0A1C7CZL1 A0A2C9GQY7 A0A182MYT3 A0A2U9B735 A0A3B4ZRT8 A0A182GFJ4 M4A2F0 A0A3B3WC23 G1ST44 A0A096M6U8 A0A087YDH9 A0A2K6FK66 A0A2K6S131 A0A1S3HTH6 A0A3B5QAV1 A0A3P8WX33 A0A1S3HS39 A0A096M429 C1C521 A0A2K6C310 A0A2K5UL24 A0A0D9RZ48 A0A096MWY6 A0A2K5NCM6 A0A2K5JWE8 F7EN38 A0A2K5Q6L9 A0A3B3VZ85 F6R171 G1QMJ8 G1KU91 A0A1Y9IVZ7 A0A182Y1H2 F6QUU0 H0V367 A0A061IL24 A0A1S2ZYJ2 A0A2J8R637 A0A3Q3M6Q7 M3W110 G3TCV5 Q96B26 G1MB91 A0A2K6FK61 A0A2K5DMW5 V4AZC8 A0A2R8ZU31 A0A2J8MLI8 A0A151IJC0 A0A384C7M9 A0A3Q7W2N8 A0A2K6S132
A0A0L7LDK9 A0A194QAD1 A0A212FHC8 I4DPA4 A0A2J7PNN7 A0A067RJ27 A0A194RGK8 A0A2P8ZAR4 D2A0W4 A0A1B6CB03 A0A1B6L851 K7J7Z8 A0A1L8DXY1 A0A158NWG7 A0A1B6FP22 A0A151I515 A0A1L8DXS8 A0A232FEW2 A0A2A3E7S5 A0A151WYP5 E2BFG5 A0A088AJ30 F4WQ24 U4U0H3 A0A026W4F8 A0A1Y1NDX8 E2A2H2 A0A182P5V7 S4RI20 A0A1Y1NDI9 A0A182JYU8 A0A0L8I2M4 K4G0P6 Q17NN4 A0A195ECK5 A0A154PGF9 A0A0L7RBM2 A0A182HBH1 D1MLN6 A0A182WZT4 Q7Q0D9 A0A1C7CZL1 A0A2C9GQY7 A0A182MYT3 A0A2U9B735 A0A3B4ZRT8 A0A182GFJ4 M4A2F0 A0A3B3WC23 G1ST44 A0A096M6U8 A0A087YDH9 A0A2K6FK66 A0A2K6S131 A0A1S3HTH6 A0A3B5QAV1 A0A3P8WX33 A0A1S3HS39 A0A096M429 C1C521 A0A2K6C310 A0A2K5UL24 A0A0D9RZ48 A0A096MWY6 A0A2K5NCM6 A0A2K5JWE8 F7EN38 A0A2K5Q6L9 A0A3B3VZ85 F6R171 G1QMJ8 G1KU91 A0A1Y9IVZ7 A0A182Y1H2 F6QUU0 H0V367 A0A061IL24 A0A1S2ZYJ2 A0A2J8R637 A0A3Q3M6Q7 M3W110 G3TCV5 Q96B26 G1MB91 A0A2K6FK61 A0A2K5DMW5 V4AZC8 A0A2R8ZU31 A0A2J8MLI8 A0A151IJC0 A0A384C7M9 A0A3Q7W2N8 A0A2K6S132
Pubmed
19121390
23622113
26354079
26227816
22118469
22651552
+ More
24845553 29403074 18362917 19820115 20075255 21347285 28648823 20798317 21719571 23537049 24508170 28004739 23056606 17510324 26483478 20920202 12364791 14747013 17210077 20966253 23542700 21993624 24487278 17431167 25319552 25243066 25244985 19892987 23929341 29704459 17975172 15057823 15489334 9466265 11719186 12419256 15231747 16912217 17545563 20531389 21269460 22814378 24989451 17174896 20010809 23254933 22722832 16136131 24813606
24845553 29403074 18362917 19820115 20075255 21347285 28648823 20798317 21719571 23537049 24508170 28004739 23056606 17510324 26483478 20920202 12364791 14747013 17210077 20966253 23542700 21993624 24487278 17431167 25319552 25243066 25244985 19892987 23929341 29704459 17975172 15057823 15489334 9466265 11719186 12419256 15231747 16912217 17545563 20531389 21269460 22814378 24989451 17174896 20010809 23254933 22722832 16136131 24813606
EMBL
BABH01000812
NWSH01000371
PCG76887.1
RSAL01000063
RVE49526.1
GAIX01006279
+ More
JAA86281.1 KQ460462 KPJ14630.1 ODYU01012731 SOQ59186.1 JTDY01001539 KOB73613.1 KQ459299 KPJ01955.1 AGBW02008504 OWR53145.1 AK403495 BAM19744.1 NEVH01023380 PNF17952.1 KK852444 KDR23812.1 KQ460205 KPJ16953.1 PYGN01000121 PSN53587.1 KQ971338 EFA02571.1 GEDC01026652 JAS10646.1 GEBQ01020099 JAT19878.1 GFDF01002832 JAV11252.1 ADTU01002971 GECZ01017827 JAS51942.1 KQ976434 KYM87187.1 GFDF01002831 JAV11253.1 NNAY01000312 OXU29296.1 KZ288338 PBC27813.1 KQ982651 KYQ52821.1 GL448037 EFN85487.1 GL888262 EGI63647.1 KB630337 ERL83540.1 KK107429 EZA50937.1 GEZM01005767 JAV95929.1 GL436038 EFN72375.1 GEZM01005765 JAV95931.1 KQ416694 KOF95713.1 JX053323 AFK11551.1 CH477198 EAT48279.1 KQ979074 KYN22841.1 KQ434899 KZC10945.1 KQ414617 KOC68332.1 JXUM01124915 KQ566885 KXJ69755.1 GU196316 ACZ26287.1 AAAB01008986 EAA00202.3 APCN01003308 CP026246 AWO99782.1 JXUM01059638 KQ562064 KXJ76762.1 AYCK01000544 AYCK01000545 BT081950 ACO52081.1 AQIA01029677 AQIB01113704 AHZZ02011068 JSUE03017956 JV046098 AFI36169.1 GAMQ01006417 GAMP01009468 JAB35434.1 JAB43287.1 ADFV01054698 ADFV01054699 AAKN02010618 KE664604 RAZU01000067 ERE89686.1 RLQ76315.1 ABGA01267752 NDHI03003746 PNJ03984.1 PNJ03985.1 AANG04001781 AL138706 CH471075 BC020773 AF025438 ACTA01100680 KB201166 ESO99081.1 AJFE02094469 AACZ04071734 NBAG03000252 PNI60372.1 PNI60373.1 KQ977362 KYN03301.1
JAA86281.1 KQ460462 KPJ14630.1 ODYU01012731 SOQ59186.1 JTDY01001539 KOB73613.1 KQ459299 KPJ01955.1 AGBW02008504 OWR53145.1 AK403495 BAM19744.1 NEVH01023380 PNF17952.1 KK852444 KDR23812.1 KQ460205 KPJ16953.1 PYGN01000121 PSN53587.1 KQ971338 EFA02571.1 GEDC01026652 JAS10646.1 GEBQ01020099 JAT19878.1 GFDF01002832 JAV11252.1 ADTU01002971 GECZ01017827 JAS51942.1 KQ976434 KYM87187.1 GFDF01002831 JAV11253.1 NNAY01000312 OXU29296.1 KZ288338 PBC27813.1 KQ982651 KYQ52821.1 GL448037 EFN85487.1 GL888262 EGI63647.1 KB630337 ERL83540.1 KK107429 EZA50937.1 GEZM01005767 JAV95929.1 GL436038 EFN72375.1 GEZM01005765 JAV95931.1 KQ416694 KOF95713.1 JX053323 AFK11551.1 CH477198 EAT48279.1 KQ979074 KYN22841.1 KQ434899 KZC10945.1 KQ414617 KOC68332.1 JXUM01124915 KQ566885 KXJ69755.1 GU196316 ACZ26287.1 AAAB01008986 EAA00202.3 APCN01003308 CP026246 AWO99782.1 JXUM01059638 KQ562064 KXJ76762.1 AYCK01000544 AYCK01000545 BT081950 ACO52081.1 AQIA01029677 AQIB01113704 AHZZ02011068 JSUE03017956 JV046098 AFI36169.1 GAMQ01006417 GAMP01009468 JAB35434.1 JAB43287.1 ADFV01054698 ADFV01054699 AAKN02010618 KE664604 RAZU01000067 ERE89686.1 RLQ76315.1 ABGA01267752 NDHI03003746 PNJ03984.1 PNJ03985.1 AANG04001781 AL138706 CH471075 BC020773 AF025438 ACTA01100680 KB201166 ESO99081.1 AJFE02094469 AACZ04071734 NBAG03000252 PNI60372.1 PNI60373.1 KQ977362 KYN03301.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000037510
UP000053268
+ More
UP000007151 UP000235965 UP000027135 UP000245037 UP000007266 UP000002358 UP000005205 UP000078540 UP000215335 UP000242457 UP000075809 UP000008237 UP000005203 UP000007755 UP000030742 UP000053097 UP000000311 UP000075885 UP000245300 UP000075881 UP000053454 UP000008820 UP000078492 UP000076502 UP000053825 UP000069940 UP000249989 UP000076407 UP000007062 UP000075882 UP000075840 UP000075884 UP000246464 UP000261400 UP000002852 UP000261480 UP000001811 UP000028760 UP000233160 UP000233220 UP000085678 UP000265120 UP000233120 UP000233100 UP000029965 UP000028761 UP000233060 UP000233080 UP000006718 UP000233040 UP000261500 UP000008225 UP000001073 UP000001646 UP000075920 UP000076408 UP000002281 UP000005447 UP000030759 UP000273346 UP000079721 UP000001595 UP000261640 UP000011712 UP000007646 UP000005640 UP000008912 UP000233020 UP000030746 UP000240080 UP000002277 UP000078542 UP000261680 UP000291021 UP000286642
UP000007151 UP000235965 UP000027135 UP000245037 UP000007266 UP000002358 UP000005205 UP000078540 UP000215335 UP000242457 UP000075809 UP000008237 UP000005203 UP000007755 UP000030742 UP000053097 UP000000311 UP000075885 UP000245300 UP000075881 UP000053454 UP000008820 UP000078492 UP000076502 UP000053825 UP000069940 UP000249989 UP000076407 UP000007062 UP000075882 UP000075840 UP000075884 UP000246464 UP000261400 UP000002852 UP000261480 UP000001811 UP000028760 UP000233160 UP000233220 UP000085678 UP000265120 UP000233120 UP000233100 UP000029965 UP000028761 UP000233060 UP000233080 UP000006718 UP000233040 UP000261500 UP000008225 UP000001073 UP000001646 UP000075920 UP000076408 UP000002281 UP000005447 UP000030759 UP000273346 UP000079721 UP000001595 UP000261640 UP000011712 UP000007646 UP000005640 UP000008912 UP000233020 UP000030746 UP000240080 UP000002277 UP000078542 UP000261680 UP000291021 UP000286642
Interpro
Gene 3D
ProteinModelPortal
H9IVA8
A0A2A4JXZ4
A0A3S2NV38
S4P3V4
A0A0N1I9Y5
A0A2H1X1V6
+ More
A0A0L7LDK9 A0A194QAD1 A0A212FHC8 I4DPA4 A0A2J7PNN7 A0A067RJ27 A0A194RGK8 A0A2P8ZAR4 D2A0W4 A0A1B6CB03 A0A1B6L851 K7J7Z8 A0A1L8DXY1 A0A158NWG7 A0A1B6FP22 A0A151I515 A0A1L8DXS8 A0A232FEW2 A0A2A3E7S5 A0A151WYP5 E2BFG5 A0A088AJ30 F4WQ24 U4U0H3 A0A026W4F8 A0A1Y1NDX8 E2A2H2 A0A182P5V7 S4RI20 A0A1Y1NDI9 A0A182JYU8 A0A0L8I2M4 K4G0P6 Q17NN4 A0A195ECK5 A0A154PGF9 A0A0L7RBM2 A0A182HBH1 D1MLN6 A0A182WZT4 Q7Q0D9 A0A1C7CZL1 A0A2C9GQY7 A0A182MYT3 A0A2U9B735 A0A3B4ZRT8 A0A182GFJ4 M4A2F0 A0A3B3WC23 G1ST44 A0A096M6U8 A0A087YDH9 A0A2K6FK66 A0A2K6S131 A0A1S3HTH6 A0A3B5QAV1 A0A3P8WX33 A0A1S3HS39 A0A096M429 C1C521 A0A2K6C310 A0A2K5UL24 A0A0D9RZ48 A0A096MWY6 A0A2K5NCM6 A0A2K5JWE8 F7EN38 A0A2K5Q6L9 A0A3B3VZ85 F6R171 G1QMJ8 G1KU91 A0A1Y9IVZ7 A0A182Y1H2 F6QUU0 H0V367 A0A061IL24 A0A1S2ZYJ2 A0A2J8R637 A0A3Q3M6Q7 M3W110 G3TCV5 Q96B26 G1MB91 A0A2K6FK61 A0A2K5DMW5 V4AZC8 A0A2R8ZU31 A0A2J8MLI8 A0A151IJC0 A0A384C7M9 A0A3Q7W2N8 A0A2K6S132
A0A0L7LDK9 A0A194QAD1 A0A212FHC8 I4DPA4 A0A2J7PNN7 A0A067RJ27 A0A194RGK8 A0A2P8ZAR4 D2A0W4 A0A1B6CB03 A0A1B6L851 K7J7Z8 A0A1L8DXY1 A0A158NWG7 A0A1B6FP22 A0A151I515 A0A1L8DXS8 A0A232FEW2 A0A2A3E7S5 A0A151WYP5 E2BFG5 A0A088AJ30 F4WQ24 U4U0H3 A0A026W4F8 A0A1Y1NDX8 E2A2H2 A0A182P5V7 S4RI20 A0A1Y1NDI9 A0A182JYU8 A0A0L8I2M4 K4G0P6 Q17NN4 A0A195ECK5 A0A154PGF9 A0A0L7RBM2 A0A182HBH1 D1MLN6 A0A182WZT4 Q7Q0D9 A0A1C7CZL1 A0A2C9GQY7 A0A182MYT3 A0A2U9B735 A0A3B4ZRT8 A0A182GFJ4 M4A2F0 A0A3B3WC23 G1ST44 A0A096M6U8 A0A087YDH9 A0A2K6FK66 A0A2K6S131 A0A1S3HTH6 A0A3B5QAV1 A0A3P8WX33 A0A1S3HS39 A0A096M429 C1C521 A0A2K6C310 A0A2K5UL24 A0A0D9RZ48 A0A096MWY6 A0A2K5NCM6 A0A2K5JWE8 F7EN38 A0A2K5Q6L9 A0A3B3VZ85 F6R171 G1QMJ8 G1KU91 A0A1Y9IVZ7 A0A182Y1H2 F6QUU0 H0V367 A0A061IL24 A0A1S2ZYJ2 A0A2J8R637 A0A3Q3M6Q7 M3W110 G3TCV5 Q96B26 G1MB91 A0A2K6FK61 A0A2K5DMW5 V4AZC8 A0A2R8ZU31 A0A2J8MLI8 A0A151IJC0 A0A384C7M9 A0A3Q7W2N8 A0A2K6S132
PDB
6H25
E-value=1.85339e-58,
Score=570
Ontologies
KEGG
GO
GO:0006401
GO:0000178
GO:0006396
GO:0004527
GO:0017091
GO:0034475
GO:0034476
GO:0000177
GO:0071042
GO:0000176
GO:0071035
GO:0016075
GO:0000467
GO:0043928
GO:0071028
GO:0034473
GO:0071038
GO:0034427
GO:0021693
GO:0007409
GO:0042552
GO:0042802
GO:0005737
GO:0035925
GO:0005634
GO:0016787
GO:0005829
GO:0006364
GO:0005654
GO:0043488
GO:0005730
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleolus
Nucleus
Nucleolus
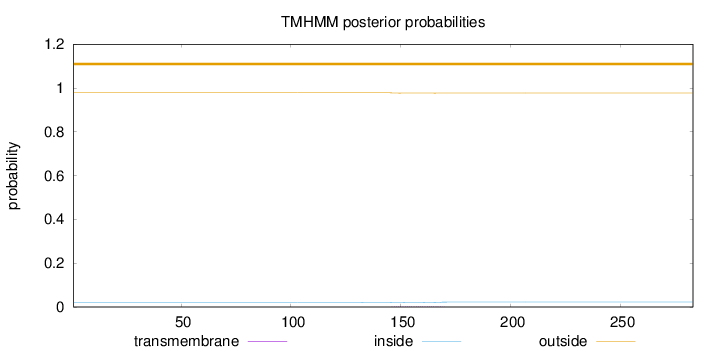
Length:
283
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05035
Exp number, first 60 AAs:
0.00041
Total prob of N-in:
0.02139
outside
1 - 283
Population Genetic Test Statistics
Pi
138.765954
Theta
133.040195
Tajima's D
-0.196703
CLR
0.583037
CSRT
0.32048397580121
Interpretation
Possibly Positive selection
