Gene
KWMTBOMO07885
Pre Gene Modal
BGIBMGA000958
Annotation
PREDICTED:_zinc_transporter_9_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 1.556
Sequence
CDS
ATGTACGACAAACGAGTCAAAGATGTAAAAACTCAGAAGATCATAAAACAGAGTCCAACTGGAGATATCATTGTTAAAACCGAAACAGGACTAGATAAAGTAATTACAAAAGTGACTATTGAAAAAAGTCAACCACAGCAAACCAAACAGGAGATGGTATCTAAGCCCGTAAAAAAAAGACTCCTAATAGATTTTTCTGCATCATACACTGAGAGAAACTTTATTACTCCCATGAGAGCTATGACTGAATACCTACTGAAACAATCAGACCTCGAGTCCTTACCAAAAGTATTAAGAAGGTCGCCATATGAATCAGAACCTCCAATAACTGTATATTATCGCAGGGATGTTGAAGCAAAAGCAATTGAGATTTGGGGTTCGAAAGAAGCATTGGAAAAAGAGTTGCTTAGAAGAGAATTAGACAGAAGAAGATATGAACAAGATGTTTTTACTGTGAAAAGAAGATTAAGAGATTACAGAAGAGAAATGAGCCATACTCGCTCAAAAGATAATGTGGAAGAAGTAGGATTAAAAACTAGATCAGGACGAGTGGTACTTACAGCTATTGGAATAAATGGATGTAATTTCTTATTTAAGCTTTGTGCCTGGTTTTACACAGGCTCACACAGTCTTTTTTCTGAATGCATCCATTCTCTTGCCGATACTGTGAACCAATTGATATTAGCGTATGGTATTCATAAGTCGGTACAGATGGCAGATCCTGACCATCCATATGGTTATACAAATATGCGATATGTCTCATCACTGATATCTGGTGTTGGTATATTCTGTGTAGGAAGTGGTTTGTCTTTCTATCACGGAGTTACAGGTATTTTGGACCCACAACCATTACATGATTTTTATTGGGCATATTTTGTACTGGGAGGCGCTGTGGTATCAGAAGGTGCCACATTAATGGTAGCATTAAGCGCCATTAGAAAAGGTGCACGAGAAGCAAATATGTCACTTTATGAATATGTAATGCGAAGCTCGGACCCTTCAGTGAATGTAGTTTTATTGGAAGATACGGCAGCTGTAGCTGGCGTGATCGTAGCAGCATCTTGTATGGCTATTTCACAATACACTGGGAATCCATTACCTGATGCCCTAGGCAGCATATTAGTCGGTACCATACTGGGTGGAGTTGCATCTTTTATTATCCTGAGTAATGTCGGTGCTTTAGTTGGCAGGTCTATACCCCACGAGCAATTGGATGAAATTAACAGCGTGTTAGAAAGAGACTTTATGATAAGAGCTATTCATGATGTGAAGGGTATAGATATAGGCAGTAACTTGATAAGATATAAGGCTGAAGTTGATTTTGATGGCAGAGCTCTCACAAGATCGTATTTAGAGAAACACGACTTGAATGCTTTACTAGAGGACATGAAGAAAATCGACACGATCGATGATGTAGAAACTTTTTTACTTAAGCATGGAGAAAATATTGTCGATATGCTTGGAGGCGAAATTGATAGAATAGAGTTAAAGCTGAGGAAAAAGTTTCCACAAATTCGGCACTGTGATTTGGAAATACTCTAA
Protein
MYDKRVKDVKTQKIIKQSPTGDIIVKTETGLDKVITKVTIEKSQPQQTKQEMVSKPVKKRLLIDFSASYTERNFITPMRAMTEYLLKQSDLESLPKVLRRSPYESEPPITVYYRRDVEAKAIEIWGSKEALEKELLRRELDRRRYEQDVFTVKRRLRDYRREMSHTRSKDNVEEVGLKTRSGRVVLTAIGINGCNFLFKLCAWFYTGSHSLFSECIHSLADTVNQLILAYGIHKSVQMADPDHPYGYTNMRYVSSLISGVGIFCVGSGLSFYHGVTGILDPQPLHDFYWAYFVLGGAVVSEGATLMVALSAIRKGAREANMSLYEYVMRSSDPSVNVVLLEDTAAVAGVIVAASCMAISQYTGNPLPDALGSILVGTILGGVASFIILSNVGALVGRSIPHEQLDEINSVLERDFMIRAIHDVKGIDIGSNLIRYKAEVDFDGRALTRSYLEKHDLNALLEDMKKIDTIDDVETFLLKHGENIVDMLGGEIDRIELKLRKKFPQIRHCDLEIL
Summary
Uniprot
A0A2A4JYL3
A0A2H1X1P8
A0A2W1BVS9
A0A3S2LAR9
A0A194Q8V4
A0A194RLU5
+ More
H9IUM8 A0A212FHD1 A0A1Y1L3E1 A0A2J7PFG3 A0A1J1IK69 A0A2J7PFH2 A0A1B6GDF3 U4UC20 A0A182TAZ3 A0A067R8D3 A0A1B6K839 A0A1B6MR51 A0A182PV61 A0A084VIL8 K7ISI9 A0A336MRB3 A0A336KXL2 E0VCC8 D2A035 N6U763 A0A2P8Z2V3 A0A182JG46 A0A182RSS6 A0A1Q3G230 A0A182YCD4 A0A182X7T2 A0A182TI06 A0A182VLG0 A0A182KTE5 A0A1Y1L5P5 A0A232EHJ9 A0A182HHX7 Q17E31 A0A1S4F6J0 A0A182NPR5 A0A182WCY9 A0A1B6DBG1 A0A1Q3G201 A0A1B6DUX7 A0A182QU65 A0A182JQT3 A0A336MLP4 A0A182MI91 A0A2M3Z8G9 E2BMF6 W5JF65 A0A2M4ALX1 A0A182FKN0 A0A2M4BH20 A0A182H538 A0A195FDC8 A0A158NBG2 A0A1I8NBB2 A0A1B0BDW5 A0A1A9Z2D0 A0A1A9V3C2 A0A0L0BM76 A0A1A9WIH0 E2AZA4 E9IR78 Q7PZR8 A0A1B0CAN3 A0A195DR96 A0A1W4X191 A0A088AUL6 A0A1I8P539 F4X8R4 E9HE24 A0A0A1X853 A0A0K8W4N8 A0A0J7KM50 A0A1L8DDJ5 W8BSY7 A0A0L7R7T5 A0A3L8DTZ6 W8BMM7 A0A2A3ET97 A0A026W6H8 A0A1B0FC30 A0A0N0BDU8 A0A151IGW8 A0A0P5BWH7 A0A0P4YRF3 A0A0P5ELX9 A0A0P5E728 A0A0P6FI84 A0A0P5SRV6 A0A0P6EAV0 A0A0P5B0Z3 A0A0P5L518 A0A0P4Y271 A0A0P5Y8N2 A0A0P4WWG8 A0A0P5UX75
H9IUM8 A0A212FHD1 A0A1Y1L3E1 A0A2J7PFG3 A0A1J1IK69 A0A2J7PFH2 A0A1B6GDF3 U4UC20 A0A182TAZ3 A0A067R8D3 A0A1B6K839 A0A1B6MR51 A0A182PV61 A0A084VIL8 K7ISI9 A0A336MRB3 A0A336KXL2 E0VCC8 D2A035 N6U763 A0A2P8Z2V3 A0A182JG46 A0A182RSS6 A0A1Q3G230 A0A182YCD4 A0A182X7T2 A0A182TI06 A0A182VLG0 A0A182KTE5 A0A1Y1L5P5 A0A232EHJ9 A0A182HHX7 Q17E31 A0A1S4F6J0 A0A182NPR5 A0A182WCY9 A0A1B6DBG1 A0A1Q3G201 A0A1B6DUX7 A0A182QU65 A0A182JQT3 A0A336MLP4 A0A182MI91 A0A2M3Z8G9 E2BMF6 W5JF65 A0A2M4ALX1 A0A182FKN0 A0A2M4BH20 A0A182H538 A0A195FDC8 A0A158NBG2 A0A1I8NBB2 A0A1B0BDW5 A0A1A9Z2D0 A0A1A9V3C2 A0A0L0BM76 A0A1A9WIH0 E2AZA4 E9IR78 Q7PZR8 A0A1B0CAN3 A0A195DR96 A0A1W4X191 A0A088AUL6 A0A1I8P539 F4X8R4 E9HE24 A0A0A1X853 A0A0K8W4N8 A0A0J7KM50 A0A1L8DDJ5 W8BSY7 A0A0L7R7T5 A0A3L8DTZ6 W8BMM7 A0A2A3ET97 A0A026W6H8 A0A1B0FC30 A0A0N0BDU8 A0A151IGW8 A0A0P5BWH7 A0A0P4YRF3 A0A0P5ELX9 A0A0P5E728 A0A0P6FI84 A0A0P5SRV6 A0A0P6EAV0 A0A0P5B0Z3 A0A0P5L518 A0A0P4Y271 A0A0P5Y8N2 A0A0P4WWG8 A0A0P5UX75
Pubmed
EMBL
NWSH01000371
PCG76899.1
ODYU01012731
SOQ59187.1
KZ149978
PZC75863.1
+ More
RSAL01000063 RVE49528.1 KQ459299 KPJ01953.1 KQ460205 KPJ16956.1 BABH01000813 BABH01000814 AGBW02008504 OWR53144.1 GEZM01067935 JAV67318.1 NEVH01025657 PNF15084.1 CVRI01000054 CRL00637.1 PNF15083.1 GECZ01009294 JAS60475.1 KB632031 ERL88156.1 KK852631 KDR19826.1 GECU01000132 JAT07575.1 GEBQ01001550 JAT38427.1 ATLV01013369 KE524854 KFB37812.1 UFQT01001659 SSX31333.1 UFQS01001035 UFQT01001035 SSX08655.1 SSX28571.1 DS235053 EEB11034.1 KQ971338 EFA01750.2 APGK01046662 KB741064 ENN74437.1 PYGN01000221 PSN50832.1 GFDL01001214 JAV33831.1 GEZM01067937 JAV67315.1 NNAY01004480 OXU17836.1 APCN01002450 CH477287 EAT44707.1 GEDC01027685 GEDC01014261 GEDC01011167 GEDC01004800 JAS09613.1 JAS23037.1 JAS26131.1 JAS32498.1 GFDL01001211 JAV33834.1 GEDC01007853 JAS29445.1 AXCN02001663 SSX31334.1 AXCM01005758 GGFM01004072 MBW24823.1 GL449209 EFN83131.1 ADMH02001585 ETN61973.1 GGFK01008472 MBW41793.1 GGFJ01003191 MBW52332.1 JXUM01110825 JXUM01110826 JXUM01110827 JXUM01110828 JXUM01110829 JXUM01110830 JXUM01110831 KQ565448 KXJ70959.1 KQ981673 KYN38207.1 ADTU01011059 JXJN01012717 JRES01001654 KNC21122.1 GL444149 EFN61247.1 GL765107 EFZ16860.1 AAAB01008986 EAA00455.3 AJWK01004122 KQ980581 KYN15435.1 GL888932 EGI57333.1 GL732626 EFX70024.1 GBXI01007459 GBXI01003736 JAD06833.1 JAD10556.1 GDHF01021919 GDHF01006282 JAI30395.1 JAI46032.1 LBMM01005665 KMQ91311.1 GFDF01009576 JAV04508.1 GAMC01006532 JAC00024.1 KQ414638 KOC66899.1 QOIP01000004 RLU23218.1 GAMC01006533 GAMC01006531 JAC00023.1 KZ288192 PBC34271.1 KK107453 EZA50619.1 CCAG010005008 KQ435850 KOX70938.1 KQ977661 KYN00667.1 GDIP01179486 JAJ43916.1 GDIP01228127 JAI95274.1 GDIP01146186 JAJ77216.1 GDIP01146184 JAJ77218.1 GDIQ01056950 JAN37787.1 GDIP01136227 LRGB01002843 JAL67487.1 KZS06165.1 GDIQ01211702 GDIQ01086456 GDIQ01066214 JAN28523.1 GDIP01191280 JAJ32122.1 GDIQ01175511 JAK76214.1 GDIP01233620 JAI89781.1 GDIP01061590 JAM42125.1 GDIP01251568 JAI71833.1 GDIP01110291 JAL93423.1
RSAL01000063 RVE49528.1 KQ459299 KPJ01953.1 KQ460205 KPJ16956.1 BABH01000813 BABH01000814 AGBW02008504 OWR53144.1 GEZM01067935 JAV67318.1 NEVH01025657 PNF15084.1 CVRI01000054 CRL00637.1 PNF15083.1 GECZ01009294 JAS60475.1 KB632031 ERL88156.1 KK852631 KDR19826.1 GECU01000132 JAT07575.1 GEBQ01001550 JAT38427.1 ATLV01013369 KE524854 KFB37812.1 UFQT01001659 SSX31333.1 UFQS01001035 UFQT01001035 SSX08655.1 SSX28571.1 DS235053 EEB11034.1 KQ971338 EFA01750.2 APGK01046662 KB741064 ENN74437.1 PYGN01000221 PSN50832.1 GFDL01001214 JAV33831.1 GEZM01067937 JAV67315.1 NNAY01004480 OXU17836.1 APCN01002450 CH477287 EAT44707.1 GEDC01027685 GEDC01014261 GEDC01011167 GEDC01004800 JAS09613.1 JAS23037.1 JAS26131.1 JAS32498.1 GFDL01001211 JAV33834.1 GEDC01007853 JAS29445.1 AXCN02001663 SSX31334.1 AXCM01005758 GGFM01004072 MBW24823.1 GL449209 EFN83131.1 ADMH02001585 ETN61973.1 GGFK01008472 MBW41793.1 GGFJ01003191 MBW52332.1 JXUM01110825 JXUM01110826 JXUM01110827 JXUM01110828 JXUM01110829 JXUM01110830 JXUM01110831 KQ565448 KXJ70959.1 KQ981673 KYN38207.1 ADTU01011059 JXJN01012717 JRES01001654 KNC21122.1 GL444149 EFN61247.1 GL765107 EFZ16860.1 AAAB01008986 EAA00455.3 AJWK01004122 KQ980581 KYN15435.1 GL888932 EGI57333.1 GL732626 EFX70024.1 GBXI01007459 GBXI01003736 JAD06833.1 JAD10556.1 GDHF01021919 GDHF01006282 JAI30395.1 JAI46032.1 LBMM01005665 KMQ91311.1 GFDF01009576 JAV04508.1 GAMC01006532 JAC00024.1 KQ414638 KOC66899.1 QOIP01000004 RLU23218.1 GAMC01006533 GAMC01006531 JAC00023.1 KZ288192 PBC34271.1 KK107453 EZA50619.1 CCAG010005008 KQ435850 KOX70938.1 KQ977661 KYN00667.1 GDIP01179486 JAJ43916.1 GDIP01228127 JAI95274.1 GDIP01146186 JAJ77216.1 GDIP01146184 JAJ77218.1 GDIQ01056950 JAN37787.1 GDIP01136227 LRGB01002843 JAL67487.1 KZS06165.1 GDIQ01211702 GDIQ01086456 GDIQ01066214 JAN28523.1 GDIP01191280 JAJ32122.1 GDIQ01175511 JAK76214.1 GDIP01233620 JAI89781.1 GDIP01061590 JAM42125.1 GDIP01251568 JAI71833.1 GDIP01110291 JAL93423.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000005204
UP000007151
+ More
UP000235965 UP000183832 UP000030742 UP000075901 UP000027135 UP000075885 UP000030765 UP000002358 UP000009046 UP000007266 UP000019118 UP000245037 UP000075880 UP000075900 UP000076408 UP000076407 UP000075902 UP000075903 UP000075882 UP000215335 UP000075840 UP000008820 UP000075884 UP000075920 UP000075886 UP000075881 UP000075883 UP000008237 UP000000673 UP000069272 UP000069940 UP000249989 UP000078541 UP000005205 UP000095301 UP000092460 UP000092445 UP000078200 UP000037069 UP000091820 UP000000311 UP000007062 UP000092461 UP000078492 UP000192223 UP000005203 UP000095300 UP000007755 UP000000305 UP000036403 UP000053825 UP000279307 UP000242457 UP000053097 UP000092444 UP000053105 UP000078542 UP000076858
UP000235965 UP000183832 UP000030742 UP000075901 UP000027135 UP000075885 UP000030765 UP000002358 UP000009046 UP000007266 UP000019118 UP000245037 UP000075880 UP000075900 UP000076408 UP000076407 UP000075902 UP000075903 UP000075882 UP000215335 UP000075840 UP000008820 UP000075884 UP000075920 UP000075886 UP000075881 UP000075883 UP000008237 UP000000673 UP000069272 UP000069940 UP000249989 UP000078541 UP000005205 UP000095301 UP000092460 UP000092445 UP000078200 UP000037069 UP000091820 UP000000311 UP000007062 UP000092461 UP000078492 UP000192223 UP000005203 UP000095300 UP000007755 UP000000305 UP000036403 UP000053825 UP000279307 UP000242457 UP000053097 UP000092444 UP000053105 UP000078542 UP000076858
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JYL3
A0A2H1X1P8
A0A2W1BVS9
A0A3S2LAR9
A0A194Q8V4
A0A194RLU5
+ More
H9IUM8 A0A212FHD1 A0A1Y1L3E1 A0A2J7PFG3 A0A1J1IK69 A0A2J7PFH2 A0A1B6GDF3 U4UC20 A0A182TAZ3 A0A067R8D3 A0A1B6K839 A0A1B6MR51 A0A182PV61 A0A084VIL8 K7ISI9 A0A336MRB3 A0A336KXL2 E0VCC8 D2A035 N6U763 A0A2P8Z2V3 A0A182JG46 A0A182RSS6 A0A1Q3G230 A0A182YCD4 A0A182X7T2 A0A182TI06 A0A182VLG0 A0A182KTE5 A0A1Y1L5P5 A0A232EHJ9 A0A182HHX7 Q17E31 A0A1S4F6J0 A0A182NPR5 A0A182WCY9 A0A1B6DBG1 A0A1Q3G201 A0A1B6DUX7 A0A182QU65 A0A182JQT3 A0A336MLP4 A0A182MI91 A0A2M3Z8G9 E2BMF6 W5JF65 A0A2M4ALX1 A0A182FKN0 A0A2M4BH20 A0A182H538 A0A195FDC8 A0A158NBG2 A0A1I8NBB2 A0A1B0BDW5 A0A1A9Z2D0 A0A1A9V3C2 A0A0L0BM76 A0A1A9WIH0 E2AZA4 E9IR78 Q7PZR8 A0A1B0CAN3 A0A195DR96 A0A1W4X191 A0A088AUL6 A0A1I8P539 F4X8R4 E9HE24 A0A0A1X853 A0A0K8W4N8 A0A0J7KM50 A0A1L8DDJ5 W8BSY7 A0A0L7R7T5 A0A3L8DTZ6 W8BMM7 A0A2A3ET97 A0A026W6H8 A0A1B0FC30 A0A0N0BDU8 A0A151IGW8 A0A0P5BWH7 A0A0P4YRF3 A0A0P5ELX9 A0A0P5E728 A0A0P6FI84 A0A0P5SRV6 A0A0P6EAV0 A0A0P5B0Z3 A0A0P5L518 A0A0P4Y271 A0A0P5Y8N2 A0A0P4WWG8 A0A0P5UX75
H9IUM8 A0A212FHD1 A0A1Y1L3E1 A0A2J7PFG3 A0A1J1IK69 A0A2J7PFH2 A0A1B6GDF3 U4UC20 A0A182TAZ3 A0A067R8D3 A0A1B6K839 A0A1B6MR51 A0A182PV61 A0A084VIL8 K7ISI9 A0A336MRB3 A0A336KXL2 E0VCC8 D2A035 N6U763 A0A2P8Z2V3 A0A182JG46 A0A182RSS6 A0A1Q3G230 A0A182YCD4 A0A182X7T2 A0A182TI06 A0A182VLG0 A0A182KTE5 A0A1Y1L5P5 A0A232EHJ9 A0A182HHX7 Q17E31 A0A1S4F6J0 A0A182NPR5 A0A182WCY9 A0A1B6DBG1 A0A1Q3G201 A0A1B6DUX7 A0A182QU65 A0A182JQT3 A0A336MLP4 A0A182MI91 A0A2M3Z8G9 E2BMF6 W5JF65 A0A2M4ALX1 A0A182FKN0 A0A2M4BH20 A0A182H538 A0A195FDC8 A0A158NBG2 A0A1I8NBB2 A0A1B0BDW5 A0A1A9Z2D0 A0A1A9V3C2 A0A0L0BM76 A0A1A9WIH0 E2AZA4 E9IR78 Q7PZR8 A0A1B0CAN3 A0A195DR96 A0A1W4X191 A0A088AUL6 A0A1I8P539 F4X8R4 E9HE24 A0A0A1X853 A0A0K8W4N8 A0A0J7KM50 A0A1L8DDJ5 W8BSY7 A0A0L7R7T5 A0A3L8DTZ6 W8BMM7 A0A2A3ET97 A0A026W6H8 A0A1B0FC30 A0A0N0BDU8 A0A151IGW8 A0A0P5BWH7 A0A0P4YRF3 A0A0P5ELX9 A0A0P5E728 A0A0P6FI84 A0A0P5SRV6 A0A0P6EAV0 A0A0P5B0Z3 A0A0P5L518 A0A0P4Y271 A0A0P5Y8N2 A0A0P4WWG8 A0A0P5UX75
PDB
2ENK
E-value=1.34749e-15,
Score=204
Ontologies
KEGG
GO
PANTHER
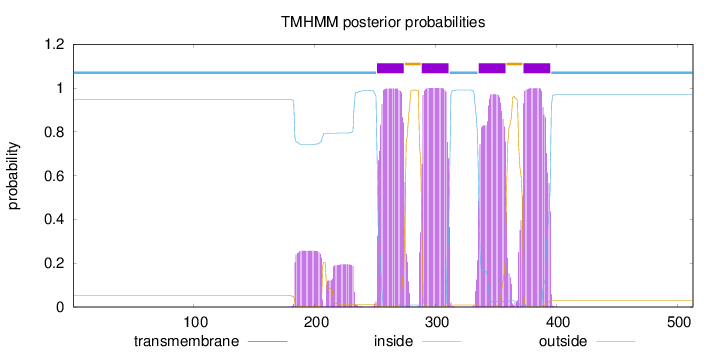
Topology
Length:
513
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
99.3101800000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.94787
inside
1 - 251
TMhelix
252 - 274
outside
275 - 288
TMhelix
289 - 311
inside
312 - 335
TMhelix
336 - 358
outside
359 - 372
TMhelix
373 - 395
inside
396 - 513
Population Genetic Test Statistics
Pi
199.187162
Theta
215.109105
Tajima's D
0.154879
CLR
0.000089
CSRT
0.410129493525324
Interpretation
Uncertain
