Pre Gene Modal
BGIBMGA001190
Annotation
PREDICTED:_uncharacterized_protein_LOC106131367_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.834
Sequence
CDS
ATGGAAAAAATGATAAATAAGAATCTCATCACCTTAAAATGCGTCCTCTTCTGTTTTCTCTCAGGGGTGGGCTGCATTTTTCCCTTTCTTCCGCTGCACATGCTTTCAGTGGGATTGGATAAAGGCGAAGCTCGTTTGATTTCTGCTGTTTCACCATGCATCGCTTTGCTAGGTCCAGCTATAGTGGGACCACTTATCGACAAATTATCAGTAGGCAGGGGATCGACGGGCGGAAGCGCTGGTAACAGCGGATCAGGTCGACTACTGCGCATCGTAACAGCAACATGCCTTATACTCAGTACTATCTTCTATTCTTTACTTCTCACTGTACCATACACGGAAAGACATGAGGCTCGTCGGCCTCAAGTTCTGTTCATGTGTAATGCGGACAGCGCGTACGTCATGCAAGAAGTTTGCAAAGAGGACATGCGTTGCAATCGTTGGTCAGGTGAGAAGTCAGGAGTACTAGCAGTTGGATCTTGTGAATACGGCTGCGCTGATCCGAACATGACTTGGATCCGCCAGCCATTCACCACCACGTCGACAACCACTACGTTAAGACCCATGTACAACACACTGGTTAACGCTTCGGCACAGACAAATGGCTCCGTTGACGAAGCCGCTCGTGAGAAAACTCACTTCGATTTAAACCCGCCTCACCTGTGTTACAACGGTGAATGTAATGTCTACATGCAACATGTTGCACGTCTGCGTATCCCGTTGTCCTTGATCGCGCCGGATCCGCCCGAGGACAACTCTACTGCTGACTCTAACTGGTGCACTTATCAGACAGCTGGTCCATCAAAGTGCTTGGTTCCGCCAGAACGATTAGCTGAGATGAACCAAGACGATGATTGCACCCCGGCAGTAAGGTGTCAAGTCCTTGACCCATACGATGAACCTGATGGTGTCTTAGCGGATGCTGAATGTAGACTTGTAATAGGAGATCCAACTTCATCATTTTGGGCTTATTTTATTCTCAGATCTATCGCAGACATATGGCCGACTGCCGCATTAGCCCTACTCAGTACAGCGTGTGTGATAGCAACTCGCGAGACCTCGTTGGGTCGAGGTGACGTTGGCAGACAAATCGCCTTCGGTACACTGGGACTGGCTATCTTCCCGCCGCTGGCCGGACTAGCCGCTGATCAAATGCCCGACACCCCATATCTTGTTCCGTTTATTCTACACGCAATCTTCATGATATTGGCTGCCTTAATATTAGTATTTGACAGTCACATGCCACTATCGACGCCGGAATGGTGGTGGCACACGGCGACAGGAGTGTTGGCCCTGCCCATGAATGTGGTGCGCCGGTATGGAGCCGAGACTGCTGCAGTTTTTGTTGTGGTAGCTCTACTGGGCATTCTGTGGAGCGGCATCGACGCATATCTTCCGTGGACCGTTGAGGAGATGAACGGCACATATACTGAAGTGGGGTTGACGCTGACCGCGGGCGCACTTCCAGCCGTGCCGGCGCTCTGGTGGGCCGAAGCACTTGTAGACTACATTGGACACTCCAATGTATTCATCTTAGCTTTTACATTTTACGGACTTCGATACACTGGTTTAGCTAGCAGTAATGAATACAGTTTGGTGGCAGTCTGTGAAATATTGGAAGTGTTTACACTTAGTTTGGTTTGGGTGACTGCAATACTATACTTCCGTCACTTGATTCCTCGCAAGTACACTACTACAGGACAAGCACTACCAGTCATCGCTCATTTCTGTTTAGGACGCTGCATTGGAGCCATTATTAGTGGGATGGTGTCGTTACAGCTGCAATCTGCCCGTGTAACGTATGAAGCCCTTGGTGTAGTTTCCTTGTTGGTAGCAGCTATATATTTGACACTCTATCATTTGTTACTTGCCCCGCGATGTGCAGCTCAAGCAGTCTCTCCGCCTCAACATTTATTACAGGGACTAAATACCAACGGAGCATCGAATGGTACATATTCTCCGATGCGGGTCTACCATGAGGAGAGATCGCGCAAAGGACACTTCCGTTACTAA
Protein
MEKMINKNLITLKCVLFCFLSGVGCIFPFLPLHMLSVGLDKGEARLISAVSPCIALLGPAIVGPLIDKLSVGRGSTGGSAGNSGSGRLLRIVTATCLILSTIFYSLLLTVPYTERHEARRPQVLFMCNADSAYVMQEVCKEDMRCNRWSGEKSGVLAVGSCEYGCADPNMTWIRQPFTTTSTTTTLRPMYNTLVNASAQTNGSVDEAAREKTHFDLNPPHLCYNGECNVYMQHVARLRIPLSLIAPDPPEDNSTADSNWCTYQTAGPSKCLVPPERLAEMNQDDDCTPAVRCQVLDPYDEPDGVLADAECRLVIGDPTSSFWAYFILRSIADIWPTAALALLSTACVIATRETSLGRGDVGRQIAFGTLGLAIFPPLAGLAADQMPDTPYLVPFILHAIFMILAALILVFDSHMPLSTPEWWWHTATGVLALPMNVVRRYGAETAAVFVVVALLGILWSGIDAYLPWTVEEMNGTYTEVGLTLTAGALPAVPALWWAEALVDYIGHSNVFILAFTFYGLRYTGLASSNEYSLVAVCEILEVFTLSLVWVTAILYFRHLIPRKYTTTGQALPVIAHFCLGRCIGAIISGMVSLQLQSARVTYEALGVVSLLVAAIYLTLYHLLLAPRCAAQAVSPPQHLLQGLNTNGASNGTYSPMRVYHEERSRKGHFRY
Summary
Uniprot
H9IVB0
A0A2A4JXZ6
S4PMZ6
A0A2H1V5X2
A0A212F5P3
A0A194Q8W1
+ More
A0A2W1BSX5 A0A194RH97 A0A0L7L1A9 E2AJP6 A0A195CMV9 F4WPB5 A0A151X3Y7 A0A195E5Q6 A0A195EV91 A0A026WJ07 A0A3L8DZ46 A0A1B0D281 A0A232ENJ0 D2A308 K7ISA3 E2BR42 A0A088A4M6 A0A0M8ZRE4 A0A0L7R355 A0A310SDZ7 A0A1Q3FL62 B0X8B8 A0A1Q3FL20 A0A1W4WK08 Q17GA0 A0A2A3EAK1 A0A182H6M6 A0A1Y1LRN3 A0A154PQ51 A0A084WJ35 A0A336M7R0 A0A182IWU6 A0A182FVT6 A0A182RR15 A0A182YL28 A0A182UT06 A0A182N3L2 A0A182UC58 A0A182L4N0 A7UVG6 A0A182HH59 A0A182W2E2 A0A182X6Q4 A0A182M1I6 A0A182KA13 A0A2J7RRX2 A0A034VUX4 A0A0A1WXK2 A0A0K8UL94 W8C4I1 A0A1A9Z3A4 A0A1A9VYP5 A0A1A9XWL6 A0A1B0B582 A0A1A9WRB7 B4J094 A0A067QW62 A0A1Q3FL95 W5JBD5 A0A1Q3FLC1 A0A0L0CR58 A0A1B6CZA8 A0A1B0FBN6 B3NFM8 B4KW74 A0A182QI49 A0A3B0K2Q0 B4HUW5 Q29DR5 B4QJS9 Q0E8H7 Q9NGV3 B4PJR1 B4LHH6 B3M9H7 Q8MQS1 U4UUC4 A0A1W4VAM1 T1PF73 A0A1I8MV67 A0A1I8QAP8 B4N2X7 B4H7A2 A0A1B6F372 A0A1B6JI39 A0A2J7RRX1 A0A1B6MRW6 A0A1B6JNX5
A0A2W1BSX5 A0A194RH97 A0A0L7L1A9 E2AJP6 A0A195CMV9 F4WPB5 A0A151X3Y7 A0A195E5Q6 A0A195EV91 A0A026WJ07 A0A3L8DZ46 A0A1B0D281 A0A232ENJ0 D2A308 K7ISA3 E2BR42 A0A088A4M6 A0A0M8ZRE4 A0A0L7R355 A0A310SDZ7 A0A1Q3FL62 B0X8B8 A0A1Q3FL20 A0A1W4WK08 Q17GA0 A0A2A3EAK1 A0A182H6M6 A0A1Y1LRN3 A0A154PQ51 A0A084WJ35 A0A336M7R0 A0A182IWU6 A0A182FVT6 A0A182RR15 A0A182YL28 A0A182UT06 A0A182N3L2 A0A182UC58 A0A182L4N0 A7UVG6 A0A182HH59 A0A182W2E2 A0A182X6Q4 A0A182M1I6 A0A182KA13 A0A2J7RRX2 A0A034VUX4 A0A0A1WXK2 A0A0K8UL94 W8C4I1 A0A1A9Z3A4 A0A1A9VYP5 A0A1A9XWL6 A0A1B0B582 A0A1A9WRB7 B4J094 A0A067QW62 A0A1Q3FL95 W5JBD5 A0A1Q3FLC1 A0A0L0CR58 A0A1B6CZA8 A0A1B0FBN6 B3NFM8 B4KW74 A0A182QI49 A0A3B0K2Q0 B4HUW5 Q29DR5 B4QJS9 Q0E8H7 Q9NGV3 B4PJR1 B4LHH6 B3M9H7 Q8MQS1 U4UUC4 A0A1W4VAM1 T1PF73 A0A1I8MV67 A0A1I8QAP8 B4N2X7 B4H7A2 A0A1B6F372 A0A1B6JI39 A0A2J7RRX1 A0A1B6MRW6 A0A1B6JNX5
Pubmed
19121390
23622113
22118469
26354079
28756777
26227816
+ More
20798317 21719571 24508170 30249741 28648823 18362917 19820115 20075255 17510324 26483478 28004739 24438588 25244985 20966253 12364791 14747013 17210077 25348373 25830018 24495485 17994087 24845553 20920257 23761445 26108605 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 23537049 25315136
20798317 21719571 24508170 30249741 28648823 18362917 19820115 20075255 17510324 26483478 28004739 24438588 25244985 20966253 12364791 14747013 17210077 25348373 25830018 24495485 17994087 24845553 20920257 23761445 26108605 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 18057021 23537049 25315136
EMBL
BABH01000821
BABH01000822
NWSH01000371
PCG76895.1
GAIX01003275
JAA89285.1
+ More
ODYU01000854 SOQ36250.1 AGBW02010161 OWR49048.1 KQ459299 KPJ01958.1 KZ149978 PZC75860.1 KQ460205 KPJ16952.1 JTDY01003662 KOB69195.1 GL440062 EFN66330.1 KQ977580 KYN01802.1 GL888243 EGI64096.1 KQ982558 KYQ54989.1 KQ979608 KYN20498.1 KQ981965 KYN31812.1 KK107182 EZA56010.1 QOIP01000003 RLU25078.1 AJVK01022592 NNAY01003136 OXU19924.1 KQ971338 EFA02250.1 GL449898 EFN81848.1 KQ435885 KOX69580.1 KQ414663 KOC65254.1 KQ760801 OAD58960.1 GFDL01006770 JAV28275.1 DS232483 EDS42454.1 GFDL01006879 JAV28166.1 CH477264 EAT45606.1 KZ288315 PBC28322.1 JXUM01114531 KQ565793 KXJ70659.1 GEZM01048872 JAV76312.1 KQ434999 KZC13378.1 ATLV01023969 KE525347 KFB50229.1 UFQS01000229 UFQT01000229 SSX01698.1 SSX22078.1 AAAB01008986 EDO63353.1 APCN01002389 AXCM01001935 NEVH01000597 PNF43577.1 GAKP01013615 GAKP01013614 GAKP01013613 JAC45338.1 GBXI01010513 GBXI01008830 GBXI01006171 JAD03779.1 JAD05462.1 JAD08121.1 GDHF01024840 JAI27474.1 GAMC01009586 JAB96969.1 JXJN01008636 JXJN01008637 CH916366 EDV95695.1 KK852872 KDR14566.1 GFDL01006763 JAV28282.1 ADMH02001988 ETN60069.1 GFDL01006733 JAV28312.1 JRES01000036 KNC34692.1 GEDC01018675 JAS18623.1 CCAG010001557 CH954178 EDV50570.1 CH933809 EDW17462.1 AXCN02001106 OUUW01000012 SPP87593.1 CH480817 EDW50736.1 CH379070 EAL30349.1 CM000363 CM002912 EDX09470.1 KMY97956.1 AE014296 BT133221 AAF50650.1 AAN12077.1 AAN12078.1 AAN12079.1 AFA28462.1 AF239609 AAF63501.1 CM000159 EDW93660.1 CH940647 EDW68506.1 CH902618 EDV41190.1 KPU78987.1 AY128419 AAM75012.1 KB632373 ERL93791.1 KA646543 AFP61172.1 CH964062 EDW78716.2 CH479217 EDW33708.1 GECZ01025131 GECZ01024977 JAS44638.1 JAS44792.1 GECU01037662 GECU01008851 JAS70044.1 JAS98855.1 PNF43576.1 GEBQ01001794 GEBQ01001319 JAT38183.1 JAT38658.1 GECU01006762 JAT00945.1
ODYU01000854 SOQ36250.1 AGBW02010161 OWR49048.1 KQ459299 KPJ01958.1 KZ149978 PZC75860.1 KQ460205 KPJ16952.1 JTDY01003662 KOB69195.1 GL440062 EFN66330.1 KQ977580 KYN01802.1 GL888243 EGI64096.1 KQ982558 KYQ54989.1 KQ979608 KYN20498.1 KQ981965 KYN31812.1 KK107182 EZA56010.1 QOIP01000003 RLU25078.1 AJVK01022592 NNAY01003136 OXU19924.1 KQ971338 EFA02250.1 GL449898 EFN81848.1 KQ435885 KOX69580.1 KQ414663 KOC65254.1 KQ760801 OAD58960.1 GFDL01006770 JAV28275.1 DS232483 EDS42454.1 GFDL01006879 JAV28166.1 CH477264 EAT45606.1 KZ288315 PBC28322.1 JXUM01114531 KQ565793 KXJ70659.1 GEZM01048872 JAV76312.1 KQ434999 KZC13378.1 ATLV01023969 KE525347 KFB50229.1 UFQS01000229 UFQT01000229 SSX01698.1 SSX22078.1 AAAB01008986 EDO63353.1 APCN01002389 AXCM01001935 NEVH01000597 PNF43577.1 GAKP01013615 GAKP01013614 GAKP01013613 JAC45338.1 GBXI01010513 GBXI01008830 GBXI01006171 JAD03779.1 JAD05462.1 JAD08121.1 GDHF01024840 JAI27474.1 GAMC01009586 JAB96969.1 JXJN01008636 JXJN01008637 CH916366 EDV95695.1 KK852872 KDR14566.1 GFDL01006763 JAV28282.1 ADMH02001988 ETN60069.1 GFDL01006733 JAV28312.1 JRES01000036 KNC34692.1 GEDC01018675 JAS18623.1 CCAG010001557 CH954178 EDV50570.1 CH933809 EDW17462.1 AXCN02001106 OUUW01000012 SPP87593.1 CH480817 EDW50736.1 CH379070 EAL30349.1 CM000363 CM002912 EDX09470.1 KMY97956.1 AE014296 BT133221 AAF50650.1 AAN12077.1 AAN12078.1 AAN12079.1 AFA28462.1 AF239609 AAF63501.1 CM000159 EDW93660.1 CH940647 EDW68506.1 CH902618 EDV41190.1 KPU78987.1 AY128419 AAM75012.1 KB632373 ERL93791.1 KA646543 AFP61172.1 CH964062 EDW78716.2 CH479217 EDW33708.1 GECZ01025131 GECZ01024977 JAS44638.1 JAS44792.1 GECU01037662 GECU01008851 JAS70044.1 JAS98855.1 PNF43576.1 GEBQ01001794 GEBQ01001319 JAT38183.1 JAT38658.1 GECU01006762 JAT00945.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000000311 UP000078542 UP000007755 UP000075809 UP000078492 UP000078541 UP000053097 UP000279307 UP000092462 UP000215335 UP000007266 UP000002358 UP000008237 UP000005203 UP000053105 UP000053825 UP000002320 UP000192223 UP000008820 UP000242457 UP000069940 UP000249989 UP000076502 UP000030765 UP000075880 UP000069272 UP000075900 UP000076408 UP000075903 UP000075884 UP000075902 UP000075882 UP000007062 UP000075840 UP000075920 UP000076407 UP000075883 UP000075881 UP000235965 UP000092445 UP000078200 UP000092443 UP000092460 UP000091820 UP000001070 UP000027135 UP000000673 UP000037069 UP000092444 UP000008711 UP000009192 UP000075886 UP000268350 UP000001292 UP000001819 UP000000304 UP000000803 UP000002282 UP000008792 UP000007801 UP000030742 UP000192221 UP000095301 UP000095300 UP000007798 UP000008744
UP000000311 UP000078542 UP000007755 UP000075809 UP000078492 UP000078541 UP000053097 UP000279307 UP000092462 UP000215335 UP000007266 UP000002358 UP000008237 UP000005203 UP000053105 UP000053825 UP000002320 UP000192223 UP000008820 UP000242457 UP000069940 UP000249989 UP000076502 UP000030765 UP000075880 UP000069272 UP000075900 UP000076408 UP000075903 UP000075884 UP000075902 UP000075882 UP000007062 UP000075840 UP000075920 UP000076407 UP000075883 UP000075881 UP000235965 UP000092445 UP000078200 UP000092443 UP000092460 UP000091820 UP000001070 UP000027135 UP000000673 UP000037069 UP000092444 UP000008711 UP000009192 UP000075886 UP000268350 UP000001292 UP000001819 UP000000304 UP000000803 UP000002282 UP000008792 UP000007801 UP000030742 UP000192221 UP000095301 UP000095300 UP000007798 UP000008744
Pfam
PF12832 MFS_1_like
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9IVB0
A0A2A4JXZ6
S4PMZ6
A0A2H1V5X2
A0A212F5P3
A0A194Q8W1
+ More
A0A2W1BSX5 A0A194RH97 A0A0L7L1A9 E2AJP6 A0A195CMV9 F4WPB5 A0A151X3Y7 A0A195E5Q6 A0A195EV91 A0A026WJ07 A0A3L8DZ46 A0A1B0D281 A0A232ENJ0 D2A308 K7ISA3 E2BR42 A0A088A4M6 A0A0M8ZRE4 A0A0L7R355 A0A310SDZ7 A0A1Q3FL62 B0X8B8 A0A1Q3FL20 A0A1W4WK08 Q17GA0 A0A2A3EAK1 A0A182H6M6 A0A1Y1LRN3 A0A154PQ51 A0A084WJ35 A0A336M7R0 A0A182IWU6 A0A182FVT6 A0A182RR15 A0A182YL28 A0A182UT06 A0A182N3L2 A0A182UC58 A0A182L4N0 A7UVG6 A0A182HH59 A0A182W2E2 A0A182X6Q4 A0A182M1I6 A0A182KA13 A0A2J7RRX2 A0A034VUX4 A0A0A1WXK2 A0A0K8UL94 W8C4I1 A0A1A9Z3A4 A0A1A9VYP5 A0A1A9XWL6 A0A1B0B582 A0A1A9WRB7 B4J094 A0A067QW62 A0A1Q3FL95 W5JBD5 A0A1Q3FLC1 A0A0L0CR58 A0A1B6CZA8 A0A1B0FBN6 B3NFM8 B4KW74 A0A182QI49 A0A3B0K2Q0 B4HUW5 Q29DR5 B4QJS9 Q0E8H7 Q9NGV3 B4PJR1 B4LHH6 B3M9H7 Q8MQS1 U4UUC4 A0A1W4VAM1 T1PF73 A0A1I8MV67 A0A1I8QAP8 B4N2X7 B4H7A2 A0A1B6F372 A0A1B6JI39 A0A2J7RRX1 A0A1B6MRW6 A0A1B6JNX5
A0A2W1BSX5 A0A194RH97 A0A0L7L1A9 E2AJP6 A0A195CMV9 F4WPB5 A0A151X3Y7 A0A195E5Q6 A0A195EV91 A0A026WJ07 A0A3L8DZ46 A0A1B0D281 A0A232ENJ0 D2A308 K7ISA3 E2BR42 A0A088A4M6 A0A0M8ZRE4 A0A0L7R355 A0A310SDZ7 A0A1Q3FL62 B0X8B8 A0A1Q3FL20 A0A1W4WK08 Q17GA0 A0A2A3EAK1 A0A182H6M6 A0A1Y1LRN3 A0A154PQ51 A0A084WJ35 A0A336M7R0 A0A182IWU6 A0A182FVT6 A0A182RR15 A0A182YL28 A0A182UT06 A0A182N3L2 A0A182UC58 A0A182L4N0 A7UVG6 A0A182HH59 A0A182W2E2 A0A182X6Q4 A0A182M1I6 A0A182KA13 A0A2J7RRX2 A0A034VUX4 A0A0A1WXK2 A0A0K8UL94 W8C4I1 A0A1A9Z3A4 A0A1A9VYP5 A0A1A9XWL6 A0A1B0B582 A0A1A9WRB7 B4J094 A0A067QW62 A0A1Q3FL95 W5JBD5 A0A1Q3FLC1 A0A0L0CR58 A0A1B6CZA8 A0A1B0FBN6 B3NFM8 B4KW74 A0A182QI49 A0A3B0K2Q0 B4HUW5 Q29DR5 B4QJS9 Q0E8H7 Q9NGV3 B4PJR1 B4LHH6 B3M9H7 Q8MQS1 U4UUC4 A0A1W4VAM1 T1PF73 A0A1I8MV67 A0A1I8QAP8 B4N2X7 B4H7A2 A0A1B6F372 A0A1B6JI39 A0A2J7RRX1 A0A1B6MRW6 A0A1B6JNX5
Ontologies
GO
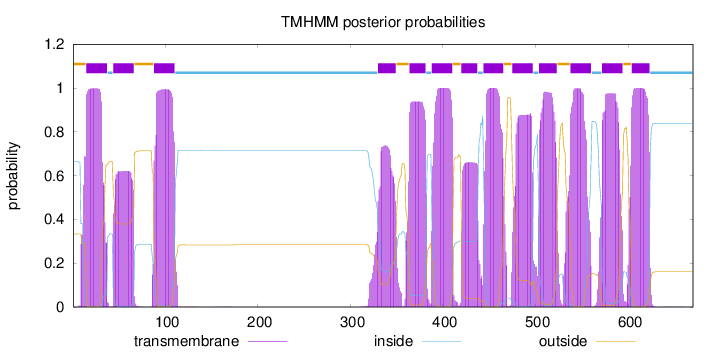
Topology
Length:
670
Number of predicted TMHs:
13
Exp number of AAs in TMHs:
252.79882
Exp number, first 60 AAs:
32.6755
Total prob of N-in:
0.66619
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 43
TMhelix
44 - 66
outside
67 - 87
TMhelix
88 - 110
inside
111 - 329
TMhelix
330 - 349
outside
350 - 363
TMhelix
364 - 381
inside
382 - 387
TMhelix
388 - 410
outside
411 - 419
TMhelix
420 - 437
inside
438 - 443
TMhelix
444 - 465
outside
466 - 474
TMhelix
475 - 497
inside
498 - 503
TMhelix
504 - 523
outside
524 - 537
TMhelix
538 - 560
inside
561 - 571
TMhelix
572 - 594
outside
595 - 603
TMhelix
604 - 623
inside
624 - 670
Population Genetic Test Statistics
Pi
236.729827
Theta
176.424784
Tajima's D
0.718199
CLR
0.567287
CSRT
0.580320983950802
Interpretation
Uncertain
