Gene
KWMTBOMO07879
Pre Gene Modal
BGIBMGA001191
Annotation
PREDICTED:_bestrophin-4-like_isoform_X2_[Bombyx_mori]
Full name
Bestrophin homolog
Location in the cell
PlasmaMembrane Reliability : 3.389
Sequence
CDS
ATGACTGTGACTTACACCGGTGAAGTAGCAACTTGCCGAGGTTTTGGGACTTTTTTAAAAGTTTTATACAGATGGCGCGGCAGTATATACAAACTGGTGTGGCTAGATCTTCTAGTTTTCCTGCTTTTGTATTATATACTCAATTTAACATATCGCTTACTTCTAGATGAAAATGCGAAGAGAACGTTCGAAGGAATAGTGAATTATTGTCATTTCCACGGGAATGTGATCCCTTTGTCGTTCGTACTAGGTTTCTATGTTACTGTAGTAATGAACCGCTGGTGGAATCAATACACAACGATACCGTGGCCTGATTCGATTGCCGTCTTCGTGTCAGCCACGATACACGGACAGGATGAACGTGGGAGGTTGATGCGTCGGACAATCGTGCGCTATGTATGTCTCTGTCTGACGATGGTGTTAACGATGATATCTCCAAGAGTCAAGAAGCGTTTTCCGACGCTCGACAACTTTACCGAGGCCGGTATAATGAATGAAAATGAAAAATCAATACTTAACAATATGAATGAAAAATTTCCAAAGACAACGAAGCACTGGTTACCAATAGTGTGGGCGGCTAGTATTGTGACGAGGGCCCGTAAGGAGGGCCGAATAAGAGACGATTTTGCCGTTAAAACTATAATAGATGAATTAAATAAATTTAGAGCACAAGCGGGACTTCTCCTCAGTTATGATACCATAAGCGTACCATTAGTTTACACTCAGGTAGTGACAATTGCTGTATACTCGTATTTCATAACGTCTGCACTCGGCAGTCAGTGGGTCGATAACAAAGTGCAGTCTGCTGAATCAACGTACATTAGTAATATTGATCTCTATTTCCCGATATTCAGTACGTTAGAATTCTTCTTCTATATGGGCTGGTTGAAAGTAGCCGAATCTCTTATAAATCCATTTGGTGAGGACGACGATGATTTCGAAGTTAACTGGATAATCGATAGAGATTTACAGGTTTCATACATGATAGTCGATGAGATGCACCATGAGCATCCCGAGTTGGTGAAAGATCAGTACTGGGACGAGGTGTTCCCGTCAGAGTTGCCGTACACGTTAGCCACGGAGAACCAGCGCGAGGAGCATCCGGAGCCTTCGACAGCTCGCGTGGCGGTGCCGGTCCGGCAGCGCAGCATGGTCTTGACCGCGCCCTCGTCTGTCAAGATCGACGAGATGGTCCCCTCCAACACCGTGAGTTACAAGTTCGCCCCGCCAGAGCAAATTCAAATGAGCGACGACGCGCAGTCTGGCATACACTTCATCGTCCACCGCGGAAGCAGTAAGCGAAAGAAGGAGGAAAGGAACTCTATGGGCTCCTCTAACTCGATGGCGTCGCTACAGCAAACGCCCATGAACAGAACTAATTCTGTCACATCGATGCTCAAAAGACTGTTCTCCAAGGAAGAGCAACGGCAACCGACTACTGCCTCTGTCACTGCCACGCAGACTGATAGCGGTAACCGTTTCACGATTTCGGCGAGCAACCCGACCTTGAACAAGCCGGCCACGGCGGGCGGCAGCATGAAGATCGCGAATGAAGTCATCGAAGAGGTCGACGAACAGGCCACCATCACGAGTATGGGCAAACGACCCGACAACAGGCCGACTGCGATGAGTTTGTTTGCGCAAAAACCTCTCCCGCCCAGTGATCCAGTTAACGTGCCGTCACGAAATTCGAATCACAGCTCCAAGGAAGACAGTCGGTTGTCCCGCTCGGCGCCGAATCAGTGGGGCGGCGAGCGCGACGACGAGCGAGACGACAAGAGTCCGACGGCGCGCTCGCTCACCGCGCGGGGCTCGTTCGCCGACATCGACGACGAGCGGCGGCCGCGTCTCGACAGCGACGTCGGCGCCTCGCTCAGCGCTGCCGGCAGCCAGACACTCATCGATGATGTGGATGGGGAGGAAGAACCCGATATAGATGACTTCAACCGGCTCAGAAGTATAAGAGAGCAGCACCGAAGAGATAAATATTTACAGAAACTAATGGCTCGGTCTGTTAGTGCTCAACAGCGCACAGGCAGTGGTGAAATATTAGACAATGATCTATTGTTAGAAGACCTACTAATCCAATCAGGAGCGTTGAAGAAAGAAGAGAACCATATAAATGGGTCAAAAGAAGACACAACTTGA
Protein
MTVTYTGEVATCRGFGTFLKVLYRWRGSIYKLVWLDLLVFLLLYYILNLTYRLLLDENAKRTFEGIVNYCHFHGNVIPLSFVLGFYVTVVMNRWWNQYTTIPWPDSIAVFVSATIHGQDERGRLMRRTIVRYVCLCLTMVLTMISPRVKKRFPTLDNFTEAGIMNENEKSILNNMNEKFPKTTKHWLPIVWAASIVTRARKEGRIRDDFAVKTIIDELNKFRAQAGLLLSYDTISVPLVYTQVVTIAVYSYFITSALGSQWVDNKVQSAESTYISNIDLYFPIFSTLEFFFYMGWLKVAESLINPFGEDDDDFEVNWIIDRDLQVSYMIVDEMHHEHPELVKDQYWDEVFPSELPYTLATENQREEHPEPSTARVAVPVRQRSMVLTAPSSVKIDEMVPSNTVSYKFAPPEQIQMSDDAQSGIHFIVHRGSSKRKKEERNSMGSSNSMASLQQTPMNRTNSVTSMLKRLFSKEEQRQPTTASVTATQTDSGNRFTISASNPTLNKPATAGGSMKIANEVIEEVDEQATITSMGKRPDNRPTAMSLFAQKPLPPSDPVNVPSRNSNHSSKEDSRLSRSAPNQWGGERDDERDDKSPTARSLTARGSFADIDDERRPRLDSDVGASLSAAGSQTLIDDVDGEEEPDIDDFNRLRSIREQHRRDKYLQKLMARSVSAQQRTGSGEILDNDLLLEDLLIQSGALKKEENHINGSKEDTT
Summary
Description
Forms chloride channels.
Similarity
Belongs to the bestrophin family.
Uniprot
I6YK90
A0A2A4JZF4
A0A2A4JXZ1
A0A2A4JZR5
I6YTA0
A0A194RHZ4
+ More
A0A2H1V5Z8 A0A194QAD6 A0A2A4JZP9 A0A212FDR1 H9IVB1 S4P8M3 A0A1J1J3H6 A0A1L8DEZ2 A0A1I8NTT4 A0A1L8DF58 A0A182RR12 A0A182W2E5 A0A182N3L5 A0A1I8NTS9 A0A182QNE1 A0A182L4L9 A0A182VLV0 A0A182P6W7 A0A182HH56 A0A1I8NTS7 A0A1I8NTS6 A0A1A9Z762 A0A1B0FEG7 A0A1A9UX44 A0A1I8NTT1 A0A1I8NTS5 A0A1A9Y607 Q7PZ58 B4LWD6 B3LW14 A0A0L0CH86 B7Z0U6 Q9V3J6 A0A0P8YJY3 A0A1I8NFW8 A0A3B0K331 A0A1I8NTR7 W5J6I9 A0A0Q9WDV2 B4K5I2 A0A2M4AKJ1 A0A0Q9WP48 A0A3B0KR61 A0A0P8ZWZ0 B4HJ86 B4PKM1 A0A0B4K651 A0A0R1E224 A0A0B4KGQ2 A0A0Q9X4D5 A0A182FVT3 A0A1W4U8Y6 A0A0Q9WXV5 A0A0P8XWV3 A0A1I8NFV5 A0A1I8NTT2 A0A1W4UK44 B4NFF2 A0A1Q3G0C8 A0A182HC67 A0A1B0ASL6 B3P4K3 A0A0Q9WPW6 A0A0Q9X9U7 A0A336LW06 A0A1W4UK78 A0A3B0K4U5 A0A1I8NFV7 A0A0Q9X4F9 A0A0R1E7N8 A0A1W4U8C5 A0A034VHS1 A0A0R1E2I5 A0A0Q9X691 Q29AK3 A0A034VEC7 A0A0Q9WW91 Q17G96 Q20C62 A0A023EVG7 A0A0A1WPS8 A0A084WJ31 A0A1I8NFW5 B4JIL3 A0A034VJD8 A0A034VGL9 A0A023F417 A0A1B0GKY5 E2ART8 A0A0K8W3D4 A0A1B6MIT8 A0A0A9XZ80 A0A146LQ26 A0A026WDM0
A0A2H1V5Z8 A0A194QAD6 A0A2A4JZP9 A0A212FDR1 H9IVB1 S4P8M3 A0A1J1J3H6 A0A1L8DEZ2 A0A1I8NTT4 A0A1L8DF58 A0A182RR12 A0A182W2E5 A0A182N3L5 A0A1I8NTS9 A0A182QNE1 A0A182L4L9 A0A182VLV0 A0A182P6W7 A0A182HH56 A0A1I8NTS7 A0A1I8NTS6 A0A1A9Z762 A0A1B0FEG7 A0A1A9UX44 A0A1I8NTT1 A0A1I8NTS5 A0A1A9Y607 Q7PZ58 B4LWD6 B3LW14 A0A0L0CH86 B7Z0U6 Q9V3J6 A0A0P8YJY3 A0A1I8NFW8 A0A3B0K331 A0A1I8NTR7 W5J6I9 A0A0Q9WDV2 B4K5I2 A0A2M4AKJ1 A0A0Q9WP48 A0A3B0KR61 A0A0P8ZWZ0 B4HJ86 B4PKM1 A0A0B4K651 A0A0R1E224 A0A0B4KGQ2 A0A0Q9X4D5 A0A182FVT3 A0A1W4U8Y6 A0A0Q9WXV5 A0A0P8XWV3 A0A1I8NFV5 A0A1I8NTT2 A0A1W4UK44 B4NFF2 A0A1Q3G0C8 A0A182HC67 A0A1B0ASL6 B3P4K3 A0A0Q9WPW6 A0A0Q9X9U7 A0A336LW06 A0A1W4UK78 A0A3B0K4U5 A0A1I8NFV7 A0A0Q9X4F9 A0A0R1E7N8 A0A1W4U8C5 A0A034VHS1 A0A0R1E2I5 A0A0Q9X691 Q29AK3 A0A034VEC7 A0A0Q9WW91 Q17G96 Q20C62 A0A023EVG7 A0A0A1WPS8 A0A084WJ31 A0A1I8NFW5 B4JIL3 A0A034VJD8 A0A034VGL9 A0A023F417 A0A1B0GKY5 E2ART8 A0A0K8W3D4 A0A1B6MIT8 A0A0A9XZ80 A0A146LQ26 A0A026WDM0
Pubmed
23300744
26354079
22118469
19121390
23622113
20966253
+ More
12364791 14747013 17210077 17994087 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 20920257 23761445 18057021 17550304 26483478 25348373 15632085 17510324 16507169 24945155 25830018 24438588 25474469 20798317 25401762 26823975 24508170
12364791 14747013 17210077 17994087 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 20920257 23761445 18057021 17550304 26483478 25348373 15632085 17510324 16507169 24945155 25830018 24438588 25474469 20798317 25401762 26823975 24508170
EMBL
JQ968534
AFN57624.1
NWSH01000371
PCG76893.1
PCG76891.1
PCG76892.1
+ More
JQ968533 AFN57623.1 KQ460205 KPJ16950.1 ODYU01000854 SOQ36248.1 KQ459299 KPJ01960.1 PCG76890.1 AGBW02009000 OWR51889.1 BABH01000826 GAIX01007212 JAA85348.1 CVRI01000069 CRL07005.1 GFDF01009068 JAV05016.1 GFDF01009067 JAV05017.1 AXCN02001106 APCN01002389 CCAG010019281 CCAG010019282 AAAB01008986 EAA00042.3 CH940650 EDW66573.1 CH902617 EDV41547.1 JRES01000409 KNC31567.1 AE014297 ACL83492.1 AF218817 AY061546 AY515709 AAF32327.1 AAF54503.1 AAL29094.1 AAR99659.1 KPU79238.1 OUUW01000013 SPP87723.1 ADMH02001988 ETN60072.1 KRF82812.1 KRF82813.1 CH933806 EDW14019.1 GGFK01007946 MBW41267.1 KRF82815.1 SPP87721.1 KPU79240.1 CH480815 EDW42758.1 CM000160 EDW96780.1 AFH06336.1 KRK03334.1 AGB95834.1 CH964251 KRF99718.1 KRG00809.1 KRG00811.1 KRG00812.1 KRG00814.1 KPU79237.1 KPU79239.1 EDW83019.1 GFDL01001813 JAV33232.1 JXUM01032902 JXUM01032903 JXUM01032904 JXUM01032905 JXUM01032906 JXUM01032907 KQ560970 KXJ80175.1 JXJN01002867 JXJN01002868 JXJN01002869 CH954181 EDV49656.1 KRF82814.1 KRG00808.1 UFQT01000184 SSX21231.1 SPP87722.1 KRG00810.1 KRK03335.1 GAKP01017295 JAC41657.1 KRK03333.1 KRF99719.1 CM000070 EAL27346.3 GAKP01017301 JAC41651.1 KRF99720.1 CH477264 EAT45610.1 DQ378293 ABD64823.1 GAPW01000596 JAC13002.1 GBXI01016038 GBXI01013233 JAC98253.1 JAD01059.1 ATLV01023968 KE525347 KFB50225.1 CH916369 EDV93094.1 GAKP01017299 JAC41653.1 GAKP01017303 JAC41649.1 GBBI01002497 JAC16215.1 AJWK01033955 AJWK01033956 AJWK01033957 AJWK01033958 GL442170 EFN63856.1 GDHF01006703 JAI45611.1 GEBQ01004110 JAT35867.1 GBHO01018370 GDHC01004583 JAG25234.1 JAQ14046.1 GDHC01008646 JAQ09983.1 KK107273 EZA53781.1
JQ968533 AFN57623.1 KQ460205 KPJ16950.1 ODYU01000854 SOQ36248.1 KQ459299 KPJ01960.1 PCG76890.1 AGBW02009000 OWR51889.1 BABH01000826 GAIX01007212 JAA85348.1 CVRI01000069 CRL07005.1 GFDF01009068 JAV05016.1 GFDF01009067 JAV05017.1 AXCN02001106 APCN01002389 CCAG010019281 CCAG010019282 AAAB01008986 EAA00042.3 CH940650 EDW66573.1 CH902617 EDV41547.1 JRES01000409 KNC31567.1 AE014297 ACL83492.1 AF218817 AY061546 AY515709 AAF32327.1 AAF54503.1 AAL29094.1 AAR99659.1 KPU79238.1 OUUW01000013 SPP87723.1 ADMH02001988 ETN60072.1 KRF82812.1 KRF82813.1 CH933806 EDW14019.1 GGFK01007946 MBW41267.1 KRF82815.1 SPP87721.1 KPU79240.1 CH480815 EDW42758.1 CM000160 EDW96780.1 AFH06336.1 KRK03334.1 AGB95834.1 CH964251 KRF99718.1 KRG00809.1 KRG00811.1 KRG00812.1 KRG00814.1 KPU79237.1 KPU79239.1 EDW83019.1 GFDL01001813 JAV33232.1 JXUM01032902 JXUM01032903 JXUM01032904 JXUM01032905 JXUM01032906 JXUM01032907 KQ560970 KXJ80175.1 JXJN01002867 JXJN01002868 JXJN01002869 CH954181 EDV49656.1 KRF82814.1 KRG00808.1 UFQT01000184 SSX21231.1 SPP87722.1 KRG00810.1 KRK03335.1 GAKP01017295 JAC41657.1 KRK03333.1 KRF99719.1 CM000070 EAL27346.3 GAKP01017301 JAC41651.1 KRF99720.1 CH477264 EAT45610.1 DQ378293 ABD64823.1 GAPW01000596 JAC13002.1 GBXI01016038 GBXI01013233 JAC98253.1 JAD01059.1 ATLV01023968 KE525347 KFB50225.1 CH916369 EDV93094.1 GAKP01017299 JAC41653.1 GAKP01017303 JAC41649.1 GBBI01002497 JAC16215.1 AJWK01033955 AJWK01033956 AJWK01033957 AJWK01033958 GL442170 EFN63856.1 GDHF01006703 JAI45611.1 GEBQ01004110 JAT35867.1 GBHO01018370 GDHC01004583 JAG25234.1 JAQ14046.1 GDHC01008646 JAQ09983.1 KK107273 EZA53781.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000005204
UP000183832
+ More
UP000095300 UP000075900 UP000075920 UP000075884 UP000075886 UP000075882 UP000075903 UP000075885 UP000075840 UP000092445 UP000092444 UP000078200 UP000092443 UP000007062 UP000008792 UP000007801 UP000037069 UP000000803 UP000095301 UP000268350 UP000000673 UP000009192 UP000001292 UP000002282 UP000007798 UP000069272 UP000192221 UP000069940 UP000249989 UP000092460 UP000008711 UP000001819 UP000008820 UP000030765 UP000001070 UP000092461 UP000000311 UP000053097
UP000095300 UP000075900 UP000075920 UP000075884 UP000075886 UP000075882 UP000075903 UP000075885 UP000075840 UP000092445 UP000092444 UP000078200 UP000092443 UP000007062 UP000008792 UP000007801 UP000037069 UP000000803 UP000095301 UP000268350 UP000000673 UP000009192 UP000001292 UP000002282 UP000007798 UP000069272 UP000192221 UP000069940 UP000249989 UP000092460 UP000008711 UP000001819 UP000008820 UP000030765 UP000001070 UP000092461 UP000000311 UP000053097
Pfam
PF01062 Bestrophin
ProteinModelPortal
I6YK90
A0A2A4JZF4
A0A2A4JXZ1
A0A2A4JZR5
I6YTA0
A0A194RHZ4
+ More
A0A2H1V5Z8 A0A194QAD6 A0A2A4JZP9 A0A212FDR1 H9IVB1 S4P8M3 A0A1J1J3H6 A0A1L8DEZ2 A0A1I8NTT4 A0A1L8DF58 A0A182RR12 A0A182W2E5 A0A182N3L5 A0A1I8NTS9 A0A182QNE1 A0A182L4L9 A0A182VLV0 A0A182P6W7 A0A182HH56 A0A1I8NTS7 A0A1I8NTS6 A0A1A9Z762 A0A1B0FEG7 A0A1A9UX44 A0A1I8NTT1 A0A1I8NTS5 A0A1A9Y607 Q7PZ58 B4LWD6 B3LW14 A0A0L0CH86 B7Z0U6 Q9V3J6 A0A0P8YJY3 A0A1I8NFW8 A0A3B0K331 A0A1I8NTR7 W5J6I9 A0A0Q9WDV2 B4K5I2 A0A2M4AKJ1 A0A0Q9WP48 A0A3B0KR61 A0A0P8ZWZ0 B4HJ86 B4PKM1 A0A0B4K651 A0A0R1E224 A0A0B4KGQ2 A0A0Q9X4D5 A0A182FVT3 A0A1W4U8Y6 A0A0Q9WXV5 A0A0P8XWV3 A0A1I8NFV5 A0A1I8NTT2 A0A1W4UK44 B4NFF2 A0A1Q3G0C8 A0A182HC67 A0A1B0ASL6 B3P4K3 A0A0Q9WPW6 A0A0Q9X9U7 A0A336LW06 A0A1W4UK78 A0A3B0K4U5 A0A1I8NFV7 A0A0Q9X4F9 A0A0R1E7N8 A0A1W4U8C5 A0A034VHS1 A0A0R1E2I5 A0A0Q9X691 Q29AK3 A0A034VEC7 A0A0Q9WW91 Q17G96 Q20C62 A0A023EVG7 A0A0A1WPS8 A0A084WJ31 A0A1I8NFW5 B4JIL3 A0A034VJD8 A0A034VGL9 A0A023F417 A0A1B0GKY5 E2ART8 A0A0K8W3D4 A0A1B6MIT8 A0A0A9XZ80 A0A146LQ26 A0A026WDM0
A0A2H1V5Z8 A0A194QAD6 A0A2A4JZP9 A0A212FDR1 H9IVB1 S4P8M3 A0A1J1J3H6 A0A1L8DEZ2 A0A1I8NTT4 A0A1L8DF58 A0A182RR12 A0A182W2E5 A0A182N3L5 A0A1I8NTS9 A0A182QNE1 A0A182L4L9 A0A182VLV0 A0A182P6W7 A0A182HH56 A0A1I8NTS7 A0A1I8NTS6 A0A1A9Z762 A0A1B0FEG7 A0A1A9UX44 A0A1I8NTT1 A0A1I8NTS5 A0A1A9Y607 Q7PZ58 B4LWD6 B3LW14 A0A0L0CH86 B7Z0U6 Q9V3J6 A0A0P8YJY3 A0A1I8NFW8 A0A3B0K331 A0A1I8NTR7 W5J6I9 A0A0Q9WDV2 B4K5I2 A0A2M4AKJ1 A0A0Q9WP48 A0A3B0KR61 A0A0P8ZWZ0 B4HJ86 B4PKM1 A0A0B4K651 A0A0R1E224 A0A0B4KGQ2 A0A0Q9X4D5 A0A182FVT3 A0A1W4U8Y6 A0A0Q9WXV5 A0A0P8XWV3 A0A1I8NFV5 A0A1I8NTT2 A0A1W4UK44 B4NFF2 A0A1Q3G0C8 A0A182HC67 A0A1B0ASL6 B3P4K3 A0A0Q9WPW6 A0A0Q9X9U7 A0A336LW06 A0A1W4UK78 A0A3B0K4U5 A0A1I8NFV7 A0A0Q9X4F9 A0A0R1E7N8 A0A1W4U8C5 A0A034VHS1 A0A0R1E2I5 A0A0Q9X691 Q29AK3 A0A034VEC7 A0A0Q9WW91 Q17G96 Q20C62 A0A023EVG7 A0A0A1WPS8 A0A084WJ31 A0A1I8NFW5 B4JIL3 A0A034VJD8 A0A034VGL9 A0A023F417 A0A1B0GKY5 E2ART8 A0A0K8W3D4 A0A1B6MIT8 A0A0A9XZ80 A0A146LQ26 A0A026WDM0
PDB
6N28
E-value=4.08996e-95,
Score=891
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell membrane
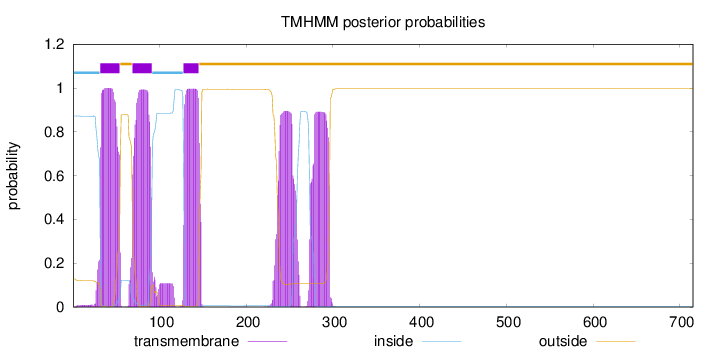
Length:
715
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
103.87683
Exp number, first 60 AAs:
22.59616
Total prob of N-in:
0.87224
POSSIBLE N-term signal
sequence
inside
1 - 31
TMhelix
32 - 54
outside
55 - 68
TMhelix
69 - 91
inside
92 - 127
TMhelix
128 - 145
outside
146 - 715
Population Genetic Test Statistics
Pi
145.448597
Theta
146.114492
Tajima's D
-0.629992
CLR
186.854393
CSRT
0.211439428028599
Interpretation
Uncertain
