Gene
KWMTBOMO07878
Pre Gene Modal
BGIBMGA001192
Annotation
PREDICTED:_uncharacterized_protein_LOC101740009_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.66
Sequence
CDS
ATGAATAAATACGCAGTGCTATTTAATTTAAAAAAAATATTAGAATATCGACCGGTTACCACAATTATAAACGCAGAAGGAAAAAATGTACCTAAAAGAAAAAACTTCGAAAACAGAATTACCTTAATTGGTACAGACAATTCAGTTAGTATAATAGATTTAAAAAATGCCCAAAATCTATCGGTAAAACGTGAATTAAAACTTGTAAAAGTTCAAGATGTAGATGTAAAAACTAGACGGCCAATTTACAAGCTGATGACTAATGCAGAATTTCACGAGGAAGAACTTGAAAGACGGAAAGAAAAAAAACTCGCTCGGCAGAACAGTTCAGTTAAAGGCCAGAAATTGATGATTTTGTCATCAAAAATATCTGAACACGATCTAATGACCAGTGTGAGAAAAATGGGAAAATTAGTAGAGAAACAGTATGAAGTTAAAGTAGTTTTAGCAGGTGAAGGAGGACAATCCTCACAAGTATTGGAAAATATATACTCAGTAATAGAGAAAAACACCAAATCAACAGCAAGATTACTACAAAAGAGAAATAAAGGTAATAGTTTAAAGTTTCAATTGCTTCCATTGAACAATGGTAACATGGAAGCATCTCAGTCAGATAATTCAGAAGATGGAAATGACAAAGGTCCGCTATGA
Protein
MNKYAVLFNLKKILEYRPVTTIINAEGKNVPKRKNFENRITLIGTDNSVSIIDLKNAQNLSVKRELKLVKVQDVDVKTRRPIYKLMTNAEFHEEELERRKEKKLARQNSSVKGQKLMILSSKISEHDLMTSVRKMGKLVEKQYEVKVVLAGEGGQSSQVLENIYSVIEKNTKSTARLLQKRNKGNSLKFQLLPLNNGNMEASQSDNSEDGNDKGPL
Summary
Similarity
Belongs to the Orn/Lys/Arg decarboxylase class-II family.
Uniprot
H9IVB2
A0A1E1WSU5
A0A2H1V649
S4P9A2
A0A212FDX7
A0A194Q8V8
+ More
A0A194RGK3 A0A2A4JYK5 A0A1E1W7Q9 A0A1S4FHU9 Q170L7 A0A182HDL7 A0A023EM14 A0A1Q3FGZ0 A0A1Q3FB53 B0WTV5 V5GLX5 W5J4K6 U4TU79 A0A1Y1KLL8 A0A182FU61 A0A2J7RFU9 A0A182JPL8 A0A182W942 A0A139WJL2 A0A182R143 A0A182VJP7 A0A182KSK8 A0A182HQ48 A0A182XNC7 A0A182MJL2 A0A182Y1R3 Q7Q084 A0A182MYL6 A0A182R5C5 A0A084WHS6 A0A182IPU9 A0A067R6I0 A0A1B0FV17 A0A0A9X1X0 A0A0A9WZW2 U5EQP9 A0A1B6KID3 A0A1J1J0J8 A0A0L0C9I1 A0A1I8NNI0 A0A1B6H431 T1GM17 A0A1B6I0Y1 A0A1B6JZ12 A0A2R7WX13 A0A336K7K6 E2AYT3 A0A1I8NA03 A0A0K8ULY5 T1PH09 A0A1A9WV25 B4MQL0 A0A034VAA8 A0A151WHZ3 C4WXA9 A0A1W4U6L5 A0A1B0ARQ6 A0A2H8TI28 A0A195FA14 A0A3L8DGQ3 A0A0C9RDF6 A0A1A9YET1 A0A232ERW9 B4KTN0 B3MCA4 E9INE1 W8BXI4 A0A3B0J5P3 A0A1A9UW88 A0A195BRD3 A0A2S2N8E6 A0A1B0G9T2 A0A026W1F9 A0A158NWU8 B4LJQ7 B4P5K5 A0A0M4EA15 B4GDN9 A0A0J7L8I0 Q292D7 B4HP87 A0A151IMS9 A0A0J9RAV7 Q4V619 B4JW55
A0A194RGK3 A0A2A4JYK5 A0A1E1W7Q9 A0A1S4FHU9 Q170L7 A0A182HDL7 A0A023EM14 A0A1Q3FGZ0 A0A1Q3FB53 B0WTV5 V5GLX5 W5J4K6 U4TU79 A0A1Y1KLL8 A0A182FU61 A0A2J7RFU9 A0A182JPL8 A0A182W942 A0A139WJL2 A0A182R143 A0A182VJP7 A0A182KSK8 A0A182HQ48 A0A182XNC7 A0A182MJL2 A0A182Y1R3 Q7Q084 A0A182MYL6 A0A182R5C5 A0A084WHS6 A0A182IPU9 A0A067R6I0 A0A1B0FV17 A0A0A9X1X0 A0A0A9WZW2 U5EQP9 A0A1B6KID3 A0A1J1J0J8 A0A0L0C9I1 A0A1I8NNI0 A0A1B6H431 T1GM17 A0A1B6I0Y1 A0A1B6JZ12 A0A2R7WX13 A0A336K7K6 E2AYT3 A0A1I8NA03 A0A0K8ULY5 T1PH09 A0A1A9WV25 B4MQL0 A0A034VAA8 A0A151WHZ3 C4WXA9 A0A1W4U6L5 A0A1B0ARQ6 A0A2H8TI28 A0A195FA14 A0A3L8DGQ3 A0A0C9RDF6 A0A1A9YET1 A0A232ERW9 B4KTN0 B3MCA4 E9INE1 W8BXI4 A0A3B0J5P3 A0A1A9UW88 A0A195BRD3 A0A2S2N8E6 A0A1B0G9T2 A0A026W1F9 A0A158NWU8 B4LJQ7 B4P5K5 A0A0M4EA15 B4GDN9 A0A0J7L8I0 Q292D7 B4HP87 A0A151IMS9 A0A0J9RAV7 Q4V619 B4JW55
Pubmed
19121390
23622113
22118469
26354079
17510324
26483478
+ More
24945155 20920257 23761445 23537049 28004739 18362917 19820115 20966253 25244985 12364791 24438588 24845553 25401762 26823975 26108605 20798317 25315136 17994087 25348373 30249741 28648823 21282665 24495485 24508170 21347285 17550304 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
24945155 20920257 23761445 23537049 28004739 18362917 19820115 20966253 25244985 12364791 24438588 24845553 25401762 26823975 26108605 20798317 25315136 17994087 25348373 30249741 28648823 21282665 24495485 24508170 21347285 17550304 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01000826
GDQN01001006
JAT90048.1
ODYU01000854
SOQ36246.1
GAIX01003829
+ More
JAA88731.1 AGBW02009000 OWR51887.1 KQ459299 KPJ01962.1 KQ460205 KPJ16948.1 NWSH01000371 PCG76889.1 GDQN01008069 JAT82985.1 CH477472 EAT40378.1 JXUM01130232 KQ567552 KXJ69377.1 GAPW01003276 JAC10322.1 GFDL01008241 JAV26804.1 GFDL01010230 JAV24815.1 DS232092 EDS34646.1 GALX01005859 JAB62607.1 ADMH02002215 ETN57749.1 KB631661 ERL85114.1 GEZM01080330 JAV62214.1 NEVH01004410 PNF39715.1 KQ971338 KYB28074.1 AXCN02001040 APCN01003292 AXCM01008879 AAAB01008986 EAA00776.4 ATLV01023877 ATLV01023878 KE525347 KFB49770.1 KK852917 KDR13865.1 AJWK01006547 GBHO01040885 GBHO01040883 GBHO01040882 GBHO01035778 GBHO01035773 GBHO01035772 GBHO01035770 GBHO01035768 GBHO01032689 GBHO01029605 GBHO01029600 GBHO01017702 GBHO01012583 GBRD01004695 GBRD01004694 GBRD01004693 JAG02719.1 JAG02721.1 JAG02722.1 JAG07826.1 JAG07831.1 JAG07832.1 JAG07834.1 JAG07836.1 JAG10915.1 JAG13999.1 JAG14004.1 JAG25902.1 JAG31021.1 JAG61126.1 GBHO01029602 GBRD01004692 GDHC01008616 JAG14002.1 JAG61129.1 JAQ10013.1 GANO01004132 JAB55739.1 GEBQ01028802 JAT11175.1 CVRI01000066 CRL05981.1 JRES01000735 KNC28902.1 GECZ01000324 JAS69445.1 CAQQ02075068 GECU01027095 JAS80611.1 GECU01003296 JAT04411.1 KK855576 PTY23175.1 UFQS01000095 UFQT01000095 SSW99355.1 SSX19735.1 GL443983 EFN61446.1 GDHF01024949 JAI27365.1 KA648051 AFP62680.1 CH963849 EDW74399.1 GAKP01019900 JAC39052.1 KQ983106 KYQ47438.1 AK342537 BAH72529.1 JXJN01002528 GFXV01001894 MBW13699.1 KQ981720 KYN37273.1 QOIP01000009 RLU19048.1 GBYB01004996 JAG74763.1 NNAY01002541 OXU21103.1 CH933808 EDW09613.1 CH902619 EDV36204.2 GL764298 EFZ17910.1 GAMC01002578 JAC03978.1 OUUW01000001 SPP75133.1 KQ976417 KYM89592.1 GGMR01000884 MBY13503.1 CCAG010007026 KK107519 EZA49421.1 ADTU01028592 CH940648 EDW60566.2 CM000158 EDW90802.1 CP012524 ALC41846.1 CH479181 EDW31696.1 LBMM01000271 KMR02225.1 CM000071 EAL24925.3 CH480816 EDW47536.1 KQ977019 KYN06259.1 CM002911 KMY93156.1 AE013599 BT022487 AAF58534.2 AAY54903.1 AHN56140.1 CH916375 EDV98193.1
JAA88731.1 AGBW02009000 OWR51887.1 KQ459299 KPJ01962.1 KQ460205 KPJ16948.1 NWSH01000371 PCG76889.1 GDQN01008069 JAT82985.1 CH477472 EAT40378.1 JXUM01130232 KQ567552 KXJ69377.1 GAPW01003276 JAC10322.1 GFDL01008241 JAV26804.1 GFDL01010230 JAV24815.1 DS232092 EDS34646.1 GALX01005859 JAB62607.1 ADMH02002215 ETN57749.1 KB631661 ERL85114.1 GEZM01080330 JAV62214.1 NEVH01004410 PNF39715.1 KQ971338 KYB28074.1 AXCN02001040 APCN01003292 AXCM01008879 AAAB01008986 EAA00776.4 ATLV01023877 ATLV01023878 KE525347 KFB49770.1 KK852917 KDR13865.1 AJWK01006547 GBHO01040885 GBHO01040883 GBHO01040882 GBHO01035778 GBHO01035773 GBHO01035772 GBHO01035770 GBHO01035768 GBHO01032689 GBHO01029605 GBHO01029600 GBHO01017702 GBHO01012583 GBRD01004695 GBRD01004694 GBRD01004693 JAG02719.1 JAG02721.1 JAG02722.1 JAG07826.1 JAG07831.1 JAG07832.1 JAG07834.1 JAG07836.1 JAG10915.1 JAG13999.1 JAG14004.1 JAG25902.1 JAG31021.1 JAG61126.1 GBHO01029602 GBRD01004692 GDHC01008616 JAG14002.1 JAG61129.1 JAQ10013.1 GANO01004132 JAB55739.1 GEBQ01028802 JAT11175.1 CVRI01000066 CRL05981.1 JRES01000735 KNC28902.1 GECZ01000324 JAS69445.1 CAQQ02075068 GECU01027095 JAS80611.1 GECU01003296 JAT04411.1 KK855576 PTY23175.1 UFQS01000095 UFQT01000095 SSW99355.1 SSX19735.1 GL443983 EFN61446.1 GDHF01024949 JAI27365.1 KA648051 AFP62680.1 CH963849 EDW74399.1 GAKP01019900 JAC39052.1 KQ983106 KYQ47438.1 AK342537 BAH72529.1 JXJN01002528 GFXV01001894 MBW13699.1 KQ981720 KYN37273.1 QOIP01000009 RLU19048.1 GBYB01004996 JAG74763.1 NNAY01002541 OXU21103.1 CH933808 EDW09613.1 CH902619 EDV36204.2 GL764298 EFZ17910.1 GAMC01002578 JAC03978.1 OUUW01000001 SPP75133.1 KQ976417 KYM89592.1 GGMR01000884 MBY13503.1 CCAG010007026 KK107519 EZA49421.1 ADTU01028592 CH940648 EDW60566.2 CM000158 EDW90802.1 CP012524 ALC41846.1 CH479181 EDW31696.1 LBMM01000271 KMR02225.1 CM000071 EAL24925.3 CH480816 EDW47536.1 KQ977019 KYN06259.1 CM002911 KMY93156.1 AE013599 BT022487 AAF58534.2 AAY54903.1 AHN56140.1 CH916375 EDV98193.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000218220
UP000008820
+ More
UP000069940 UP000249989 UP000002320 UP000000673 UP000030742 UP000069272 UP000235965 UP000075881 UP000075920 UP000007266 UP000075886 UP000075903 UP000075882 UP000075840 UP000076407 UP000075883 UP000076408 UP000007062 UP000075884 UP000075900 UP000030765 UP000075880 UP000027135 UP000092461 UP000183832 UP000037069 UP000095300 UP000015102 UP000000311 UP000095301 UP000091820 UP000007798 UP000075809 UP000192221 UP000092460 UP000078541 UP000279307 UP000092443 UP000215335 UP000009192 UP000007801 UP000268350 UP000078200 UP000078540 UP000092444 UP000053097 UP000005205 UP000008792 UP000002282 UP000092553 UP000008744 UP000036403 UP000001819 UP000001292 UP000078542 UP000000803 UP000001070
UP000069940 UP000249989 UP000002320 UP000000673 UP000030742 UP000069272 UP000235965 UP000075881 UP000075920 UP000007266 UP000075886 UP000075903 UP000075882 UP000075840 UP000076407 UP000075883 UP000076408 UP000007062 UP000075884 UP000075900 UP000030765 UP000075880 UP000027135 UP000092461 UP000183832 UP000037069 UP000095300 UP000015102 UP000000311 UP000095301 UP000091820 UP000007798 UP000075809 UP000192221 UP000092460 UP000078541 UP000279307 UP000092443 UP000215335 UP000009192 UP000007801 UP000268350 UP000078200 UP000078540 UP000092444 UP000053097 UP000005205 UP000008792 UP000002282 UP000092553 UP000008744 UP000036403 UP000001819 UP000001292 UP000078542 UP000000803 UP000001070
Pfam
Interpro
IPR001288
Translation_initiation_fac_3
+ More
IPR036788 T_IF-3_C_sf
IPR019814 Translation_initiation_fac_3_N
IPR036787 T_IF-3_N_sf
IPR000183 Orn/DAP/Arg_de-COase
IPR022657 De-COase2_CS
IPR022644 De-COase2_N
IPR009006 Ala_racemase/Decarboxylase_C
IPR002433 Orn_de-COase
IPR022653 De-COase2_pyr-phos_BS
IPR022643 De-COase2_C
IPR029066 PLP-binding_barrel
IPR000239 GPCR_kinase
IPR000719 Prot_kinase_dom
IPR036305 RGS_sf
IPR038545 Znf_DBF_sf
IPR000961 AGC-kinase_C
IPR001849 PH_domain
IPR011009 Kinase-like_dom_sf
IPR016137 RGS
IPR008271 Ser/Thr_kinase_AS
IPR006572 Znf_DBF
IPR017441 Protein_kinase_ATP_BS
IPR011993 PH-like_dom_sf
IPR019815 Translation_initiation_fac_3_C
IPR026705 Hid-1/Ecm30
IPR036788 T_IF-3_C_sf
IPR019814 Translation_initiation_fac_3_N
IPR036787 T_IF-3_N_sf
IPR000183 Orn/DAP/Arg_de-COase
IPR022657 De-COase2_CS
IPR022644 De-COase2_N
IPR009006 Ala_racemase/Decarboxylase_C
IPR002433 Orn_de-COase
IPR022653 De-COase2_pyr-phos_BS
IPR022643 De-COase2_C
IPR029066 PLP-binding_barrel
IPR000239 GPCR_kinase
IPR000719 Prot_kinase_dom
IPR036305 RGS_sf
IPR038545 Znf_DBF_sf
IPR000961 AGC-kinase_C
IPR001849 PH_domain
IPR011009 Kinase-like_dom_sf
IPR016137 RGS
IPR008271 Ser/Thr_kinase_AS
IPR006572 Znf_DBF
IPR017441 Protein_kinase_ATP_BS
IPR011993 PH-like_dom_sf
IPR019815 Translation_initiation_fac_3_C
IPR026705 Hid-1/Ecm30
SUPFAM
ProteinModelPortal
H9IVB2
A0A1E1WSU5
A0A2H1V649
S4P9A2
A0A212FDX7
A0A194Q8V8
+ More
A0A194RGK3 A0A2A4JYK5 A0A1E1W7Q9 A0A1S4FHU9 Q170L7 A0A182HDL7 A0A023EM14 A0A1Q3FGZ0 A0A1Q3FB53 B0WTV5 V5GLX5 W5J4K6 U4TU79 A0A1Y1KLL8 A0A182FU61 A0A2J7RFU9 A0A182JPL8 A0A182W942 A0A139WJL2 A0A182R143 A0A182VJP7 A0A182KSK8 A0A182HQ48 A0A182XNC7 A0A182MJL2 A0A182Y1R3 Q7Q084 A0A182MYL6 A0A182R5C5 A0A084WHS6 A0A182IPU9 A0A067R6I0 A0A1B0FV17 A0A0A9X1X0 A0A0A9WZW2 U5EQP9 A0A1B6KID3 A0A1J1J0J8 A0A0L0C9I1 A0A1I8NNI0 A0A1B6H431 T1GM17 A0A1B6I0Y1 A0A1B6JZ12 A0A2R7WX13 A0A336K7K6 E2AYT3 A0A1I8NA03 A0A0K8ULY5 T1PH09 A0A1A9WV25 B4MQL0 A0A034VAA8 A0A151WHZ3 C4WXA9 A0A1W4U6L5 A0A1B0ARQ6 A0A2H8TI28 A0A195FA14 A0A3L8DGQ3 A0A0C9RDF6 A0A1A9YET1 A0A232ERW9 B4KTN0 B3MCA4 E9INE1 W8BXI4 A0A3B0J5P3 A0A1A9UW88 A0A195BRD3 A0A2S2N8E6 A0A1B0G9T2 A0A026W1F9 A0A158NWU8 B4LJQ7 B4P5K5 A0A0M4EA15 B4GDN9 A0A0J7L8I0 Q292D7 B4HP87 A0A151IMS9 A0A0J9RAV7 Q4V619 B4JW55
A0A194RGK3 A0A2A4JYK5 A0A1E1W7Q9 A0A1S4FHU9 Q170L7 A0A182HDL7 A0A023EM14 A0A1Q3FGZ0 A0A1Q3FB53 B0WTV5 V5GLX5 W5J4K6 U4TU79 A0A1Y1KLL8 A0A182FU61 A0A2J7RFU9 A0A182JPL8 A0A182W942 A0A139WJL2 A0A182R143 A0A182VJP7 A0A182KSK8 A0A182HQ48 A0A182XNC7 A0A182MJL2 A0A182Y1R3 Q7Q084 A0A182MYL6 A0A182R5C5 A0A084WHS6 A0A182IPU9 A0A067R6I0 A0A1B0FV17 A0A0A9X1X0 A0A0A9WZW2 U5EQP9 A0A1B6KID3 A0A1J1J0J8 A0A0L0C9I1 A0A1I8NNI0 A0A1B6H431 T1GM17 A0A1B6I0Y1 A0A1B6JZ12 A0A2R7WX13 A0A336K7K6 E2AYT3 A0A1I8NA03 A0A0K8ULY5 T1PH09 A0A1A9WV25 B4MQL0 A0A034VAA8 A0A151WHZ3 C4WXA9 A0A1W4U6L5 A0A1B0ARQ6 A0A2H8TI28 A0A195FA14 A0A3L8DGQ3 A0A0C9RDF6 A0A1A9YET1 A0A232ERW9 B4KTN0 B3MCA4 E9INE1 W8BXI4 A0A3B0J5P3 A0A1A9UW88 A0A195BRD3 A0A2S2N8E6 A0A1B0G9T2 A0A026W1F9 A0A158NWU8 B4LJQ7 B4P5K5 A0A0M4EA15 B4GDN9 A0A0J7L8I0 Q292D7 B4HP87 A0A151IMS9 A0A0J9RAV7 Q4V619 B4JW55
Ontologies
GO
Topology
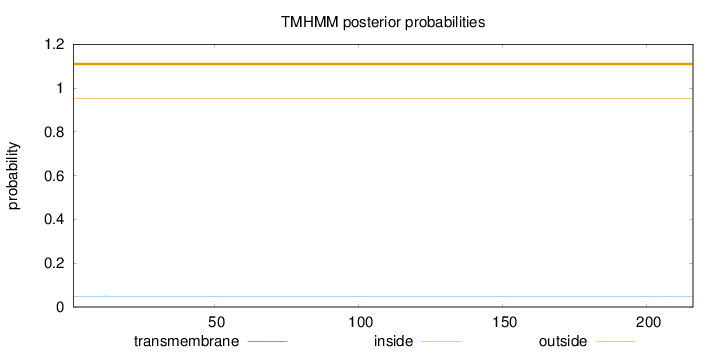
Length:
216
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00051
Exp number, first 60 AAs:
0.00051
Total prob of N-in:
0.04945
outside
1 - 216
Population Genetic Test Statistics
Pi
5.839821
Theta
43.014353
Tajima's D
-2.381177
CLR
7250.594549
CSRT
0.00189990500474976
Interpretation
Possibly Positive selection
