Pre Gene Modal
BGIBMGA000950
Annotation
PREDICTED:_THO_complex_subunit_2_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.02 Nuclear Reliability : 1.504
Sequence
CDS
ATGTCTGGATTTTCTCAAAGTGTCTATGATTTTCTTCTTTGTGGCCTGAAAGGAAACCTTAAAAAAGATACCGTTTTATCCTTCTTAAGAGAAATAACTAATTTGCATGCAGATGTGCCATCAATAATATTGGATATCATATCAATTTTGGATGCTGAAACCAGCCTAGATGTACAAAGCGATGAGCGGGCTTATTTCTGCTACATTGTGAGAGAGCTGGAGCCATTTATTTCTGATAAGTTACTAAAAGAAAGATTAGAGATAGATACATTACAAGATGCTGGTACTCTTAAGAACAAAAACTTCTACACCAAATTCATAAAGATCAAGACTAAATTATACTACAAACAACGTAAATTTAACTTATTTCGAGAAGAAAGTGAAGGATACTCAAAGTTAATAGTTGAATTAAACCAAGAAATAAATGAAGATACAGAATGGAAAAATTTGCTTGAAATAATTCAATCTCTTATTGGTTGTTTTAATTTGGATCCAAACAGGGTACTTGACATAATTCTAGAATCGTTTGAAGCCCGACCACAATTAGATGCTCTTTTTATACCGCTTATTAAGAGTTACATGGCAGATCCTCATGTAATTTCTGAGGTTTTAGGTTTCAAACTGAGTAATATGGAAATTCTTGATAACTATACAGAACCTCCACCGTTGATGACTGTAATTGCATTGCTTTTACAACATCAGGTTATATCATTAGATGACATCTATCCATGGCTTCGCCCAGATGATGTGATAATAGCTAAACAAGCTGATAAAGAAATAAAAGCAGTTCAAGAATATATAAGAAAAATTAACATTGTGTCTACCAAAGGACCACAAACAAATGCCCCCACTGAATTTATAGAAGATAAAGCTGATCCGCAGGAATATTGGAGCAATCAAAAGCTAGTGTTATGTGAAACTTTATTGAGAGTCAATGCGTGGCGAGAGTTTGCCTCACTGTTTGCGCGTCTTCCCACTACTACGATGCCGCCAAGACCAGCAATAGCCCTTTGTGGGTTGCTGCATGCTGTTGTAGAACCATTGTATCGAAAGCAATGCCGAGTAGCTTCAAAAATACTCGGAAAACCAGTACCTCCACTTAAATCAAATTTAGCACCACCTAATTGTAAAACATTCGAAGACATGAAAGAAACAGTTATACCAGCCCTTATATTGCTTGGTCCAGCCCTTCATCATGACCCTGTCTTGATGTACAAAGTCATTAGAATACTTAGAACTGCTCGCGGTCAAAAAGATGATCCGTTACATTTTGAGGCACTAACCGTTTTGGATACCGCAATATTACCGGCTCTTACACTGATGGAAGGGAATTGTTGTATGGCTGAAGAGGTGTACACATTACTTAAGTTATATCCATATCAGTGCAGATACTGTCTCTATGCGCGTTGGAAAAATGAGGCTGGCGAACGAATACCTGCTCTCATTCGTGTCCGGGGGAACTCATTGCAGAAAATAAAACACATAATGAAACGAGTATCCAAAGAGAATATCAAGCCCCAAGGTCGTCTCATCGGGAAACTGTCGCACGCCGCCCCGGCCTTTCTGTTTGATTACGTGCTCTTGCAGATACAAACATACGACAATCTGATAGGACCAGTAGTAGAATCCATGAAATATTTGACGTCTCTATCTTTAGATGTTCTGGGATACTGTTTAGTTGAGGCTTTAGGCTCTCGACGTTCGACGCCACCTGCGTGGCTACAAGCACTAGCCTCGTTCGCCGGGGCAGCCTTCAAGAAACACAATATTGAGTTAACTGCTCTTTTGCAGTTTGTTGCTAATCGACTCAAAGCTCAACAGAGTCAAGACCTGATAGTATTAAAAGAAATAGTACAAAAAATGGCCGGTATAGAAGCTGCTGAGGAAATGACACCCGAGCAGTTAGAGGCTATGGCAGGAGGTGAACTACTTAAAGGAGAGGCTGGTTACTTTTCACAAGTGCGTAACACGAGACGCTCTTCTGCGCGACTGAAAGATGCCATAGTCGGAAACAACTTAGACATATCTCTTTGTATATTGGCGGCGCAACAGAGACATTGTTGTGTTTGGAAAGAATACGAAGATGACATTCAAATAAGTGGCGAACCTCCAGGGTCCCAGCTAAAGGTGGTGGGTAGATTGGCAGACCAATGTCAGGATGCTCTGGTTCAGCTTGGTACATTCTTAGCATCGTCACATGCTCCCGATGAATATGCGGCGAGGCTGCCATCGTTACAGGAACTGCTGCGTGATTACCATGTAGATGCAGATGTCGCATTCTTTTTGCATCGTCCAGTTATAAACCAAAAGATATCCGCCAAGTTGGAATCTTTACGTAAAGCATCGGACAGCAAATCGGATTCAATAGAGAAGATTATAGAAAGATACAATGTGGCATCAAAGGAAGCCTTGGAGCCTATTGTATCGTCGATACAACCGATGCTGCCTGCGAGGGTTTTAGAAGACATTTCGCCTGAGTTTTACGTTACATTCTGGTCATTATCCATGTACGATCTTCGAGTGCCGGTCGAAAGTTACGAGAGGGAAATAGAACGCTTAAAAACAGCAGCAGCTAACGTCAGCAAGGATTCGTCTCAAGGAACCAAAGTTAAGAAAGAACAGGAACGTTTCAATACGCTCATCGAAAAACTACAGGAGGAGCGGCGCCGACAGGCCGCGCACGTGGCGGCGGTGCGGGCGCGGCTGGCGCGCGAGTGCGAGGACTGGTTCCCGGCGCGGGCGGTCCGCTCCGCCAAGAACGAGACCGTGACGCGGCTCATGCAGCTGTGCATATTCCCGCGCTGCGTGTTCACGGCGGCCGATGCCCTCTACTGCGCCGAGTTCGTGCATACCGTGCACTCTCTGAAGACTCCCAATTTCTCCACTCTTCTGTGTTATGATAGACTAAATGAATTTATAAATTTGAATTTGAAATTCTTAACCAAAGAGGAGATATTACAGATCAAAGTGATCAATACGAAAAAACGATAA
Protein
MSGFSQSVYDFLLCGLKGNLKKDTVLSFLREITNLHADVPSIILDIISILDAETSLDVQSDERAYFCYIVRELEPFISDKLLKERLEIDTLQDAGTLKNKNFYTKFIKIKTKLYYKQRKFNLFREESEGYSKLIVELNQEINEDTEWKNLLEIIQSLIGCFNLDPNRVLDIILESFEARPQLDALFIPLIKSYMADPHVISEVLGFKLSNMEILDNYTEPPPLMTVIALLLQHQVISLDDIYPWLRPDDVIIAKQADKEIKAVQEYIRKINIVSTKGPQTNAPTEFIEDKADPQEYWSNQKLVLCETLLRVNAWREFASLFARLPTTTMPPRPAIALCGLLHAVVEPLYRKQCRVASKILGKPVPPLKSNLAPPNCKTFEDMKETVIPALILLGPALHHDPVLMYKVIRILRTARGQKDDPLHFEALTVLDTAILPALTLMEGNCCMAEEVYTLLKLYPYQCRYCLYARWKNEAGERIPALIRVRGNSLQKIKHIMKRVSKENIKPQGRLIGKLSHAAPAFLFDYVLLQIQTYDNLIGPVVESMKYLTSLSLDVLGYCLVEALGSRRSTPPAWLQALASFAGAAFKKHNIELTALLQFVANRLKAQQSQDLIVLKEIVQKMAGIEAAEEMTPEQLEAMAGGELLKGEAGYFSQVRNTRRSSARLKDAIVGNNLDISLCILAAQQRHCCVWKEYEDDIQISGEPPGSQLKVVGRLADQCQDALVQLGTFLASSHAPDEYAARLPSLQELLRDYHVDADVAFFLHRPVINQKISAKLESLRKASDSKSDSIEKIIERYNVASKEALEPIVSSIQPMLPARVLEDISPEFYVTFWSLSMYDLRVPVESYEREIERLKTAAANVSKDSSQGTKVKKEQERFNTLIEKLQEERRRQAAHVAAVRARLARECEDWFPARAVRSAKNETVTRLMQLCIFPRCVFTAADALYCAEFVHTVHSLKTPNFSTLLCYDRLNEFINLNLKFLTKEEILQIKVINTKKR
Summary
Uniprot
H9IUM0
A0A212F4H1
A0A194QES9
A0A2A4JTU6
A0A2A4JU20
A0A2H1X5U1
+ More
S4P782 A0A067R8H4 A0A2J7QS75 A0A2J7QS78 A0A310SSF8 E2BUW2 E2AKU8 A0A026WUF4 A0A3L8DEF8 A0A0L7RAQ1 A0A151XJF8 A0A195BFF3 A0A195C2R0 A0A2A3E5I4 A0A154NXX1 A0A088ASY0 A0A151JQB9 K7J2Y9 Q17KE9 B0XJB9 W4VRJ6 A0A182GHW6 A0A084W4P6 A0A182MYW7 A0A182J8B0 A0A182Y3K0 A0A182R0W0 A0A182PCL3 A0A182K3M7 A0A182M4N4 A0A182W6T4 A0A182TF23 A0A182HUD7 A0A2Y9D1F1 A0A182XL18 A0A1S4GYF1 A0A182UPQ9 A0A1Q3F986 A0A1Q3F900 Q7Q461 W5JVX1 D2A126 A0A2M4A3D7 A0A2M4A3M1 A0A2M4A6A5 A0A2M4B8U5 A0A2M4B9F4 A0A0K8U3Y2 A0A0M9A0Q5 A0A0K8U870 W8BMU3 A0A182F152 A0A0K8UY07 A0A1Y1N264 A0A0A1WN99 A0A0L0C6D2 A0A1I8Q9L8 A0A1A9WZI2 A0A1A9ZDA5 A0A1B0C1B0 A0A1A9Y4B4 A0A1A9VDT8 A0A1A9YUH8 A0A1I8N9W6 A0A1J1HMU4 E0VDI7 A0A1J1HL10 A0A336K4B9 W8AY15 A0A0K8U8A5 A0A0K8UJP9 A0A1B6BZM5 A0A2M3Z0N7 A0A0K8TW02 U4UDC5 A0A182R559 A0A0P5DI62 A0A0P5YFK4 A0A0P5URC7 A0A0P5UTS9 A0A0P5E7X3 A0A0P5CNV2 A0A0N8DGI9 A0A0P6INA4 A0A0P5ZVR3 A0A0N8ACM8 A0A0P5CV75 A0A0P6DBC2 A0A0P5CUX4 A0A0P5WPY5 A0A1B6C5X2 A0A0P5NIE0 A0A0P5DXT6 A0A0P4Y4W4
S4P782 A0A067R8H4 A0A2J7QS75 A0A2J7QS78 A0A310SSF8 E2BUW2 E2AKU8 A0A026WUF4 A0A3L8DEF8 A0A0L7RAQ1 A0A151XJF8 A0A195BFF3 A0A195C2R0 A0A2A3E5I4 A0A154NXX1 A0A088ASY0 A0A151JQB9 K7J2Y9 Q17KE9 B0XJB9 W4VRJ6 A0A182GHW6 A0A084W4P6 A0A182MYW7 A0A182J8B0 A0A182Y3K0 A0A182R0W0 A0A182PCL3 A0A182K3M7 A0A182M4N4 A0A182W6T4 A0A182TF23 A0A182HUD7 A0A2Y9D1F1 A0A182XL18 A0A1S4GYF1 A0A182UPQ9 A0A1Q3F986 A0A1Q3F900 Q7Q461 W5JVX1 D2A126 A0A2M4A3D7 A0A2M4A3M1 A0A2M4A6A5 A0A2M4B8U5 A0A2M4B9F4 A0A0K8U3Y2 A0A0M9A0Q5 A0A0K8U870 W8BMU3 A0A182F152 A0A0K8UY07 A0A1Y1N264 A0A0A1WN99 A0A0L0C6D2 A0A1I8Q9L8 A0A1A9WZI2 A0A1A9ZDA5 A0A1B0C1B0 A0A1A9Y4B4 A0A1A9VDT8 A0A1A9YUH8 A0A1I8N9W6 A0A1J1HMU4 E0VDI7 A0A1J1HL10 A0A336K4B9 W8AY15 A0A0K8U8A5 A0A0K8UJP9 A0A1B6BZM5 A0A2M3Z0N7 A0A0K8TW02 U4UDC5 A0A182R559 A0A0P5DI62 A0A0P5YFK4 A0A0P5URC7 A0A0P5UTS9 A0A0P5E7X3 A0A0P5CNV2 A0A0N8DGI9 A0A0P6INA4 A0A0P5ZVR3 A0A0N8ACM8 A0A0P5CV75 A0A0P6DBC2 A0A0P5CUX4 A0A0P5WPY5 A0A1B6C5X2 A0A0P5NIE0 A0A0P5DXT6 A0A0P4Y4W4
Pubmed
EMBL
BABH01000840
BABH01000841
AGBW02010374
OWR48624.1
KQ459299
KPJ01971.1
+ More
NWSH01000673 PCG74893.1 PCG74892.1 ODYU01013259 SOQ59964.1 GAIX01004729 JAA87831.1 KK852866 KDR14733.1 NEVH01011878 PNF31425.1 PNF31427.1 KQ760498 OAD60086.1 GL450772 EFN80499.1 GL440413 EFN65937.1 KK107119 EZA58739.1 QOIP01000009 RLU18825.1 KQ414618 KOC67992.1 KQ982068 KYQ60556.1 KQ976491 KYM83306.1 KQ978379 KYM94496.1 KZ288361 PBC26997.1 KQ434781 KZC04463.1 KQ978646 KYN29434.1 CH477225 EAT47141.1 EAT47142.1 DS233478 EDS30156.1 GANO01002319 JAB57552.1 JXUM01012233 KQ560349 KXJ82903.1 ATLV01020364 KE525299 KFB45190.1 AXCN02000817 AXCM01006213 APCN01002453 AAAB01008964 GFDL01010943 JAV24102.1 GFDL01010990 JAV24055.1 EAA12368.4 ADMH02000146 ETN67613.1 KQ971338 EFA01592.2 GGFK01001940 MBW35261.1 GGFK01002024 MBW35345.1 GGFK01002837 MBW36158.1 GGFJ01000312 MBW49453.1 GGFJ01000317 MBW49458.1 GDHF01030870 JAI21444.1 KQ435798 KOX73449.1 GDHF01029457 GDHF01025770 GDHF01020633 GDHF01019694 JAI22857.1 JAI26544.1 JAI31681.1 JAI32620.1 GAMC01015591 JAB90964.1 GDHF01020765 JAI31549.1 GEZM01019409 JAV89647.1 GBXI01014414 JAC99877.1 JRES01000842 KNC27807.1 JXJN01023964 CCAG010009523 CVRI01000010 CRK88716.1 DS235078 EEB11443.1 CRK88717.1 UFQS01000045 UFQT01000045 SSW98418.1 SSX18804.1 GAMC01015588 JAB90967.1 GDHF01029422 GDHF01027328 GDHF01025816 JAI22892.1 JAI24986.1 JAI26498.1 GDHF01025581 JAI26733.1 GEDC01030829 JAS06469.1 GGFM01001332 MBW22083.1 GDHF01033858 JAI18456.1 KB632081 ERL88596.1 GDIP01157117 JAJ66285.1 GDIP01060232 JAM43483.1 GDIP01111275 JAL92439.1 GDIP01125522 JAL78192.1 GDIP01163717 JAJ59685.1 GDIP01171560 JAJ51842.1 GDIP01035935 JAM67780.1 GDIQ01029049 JAN65688.1 GDIP01038384 JAM65331.1 GDIP01160513 JAJ62889.1 GDIP01165912 JAJ57490.1 GDIQ01079777 JAN14960.1 GDIP01165213 JAJ58189.1 GDIP01083233 JAM20482.1 GEDC01028396 JAS08902.1 GDIQ01145714 GDIQ01143911 JAL06012.1 GDIP01155667 JAJ67735.1 GDIP01234762 JAI88639.1
NWSH01000673 PCG74893.1 PCG74892.1 ODYU01013259 SOQ59964.1 GAIX01004729 JAA87831.1 KK852866 KDR14733.1 NEVH01011878 PNF31425.1 PNF31427.1 KQ760498 OAD60086.1 GL450772 EFN80499.1 GL440413 EFN65937.1 KK107119 EZA58739.1 QOIP01000009 RLU18825.1 KQ414618 KOC67992.1 KQ982068 KYQ60556.1 KQ976491 KYM83306.1 KQ978379 KYM94496.1 KZ288361 PBC26997.1 KQ434781 KZC04463.1 KQ978646 KYN29434.1 CH477225 EAT47141.1 EAT47142.1 DS233478 EDS30156.1 GANO01002319 JAB57552.1 JXUM01012233 KQ560349 KXJ82903.1 ATLV01020364 KE525299 KFB45190.1 AXCN02000817 AXCM01006213 APCN01002453 AAAB01008964 GFDL01010943 JAV24102.1 GFDL01010990 JAV24055.1 EAA12368.4 ADMH02000146 ETN67613.1 KQ971338 EFA01592.2 GGFK01001940 MBW35261.1 GGFK01002024 MBW35345.1 GGFK01002837 MBW36158.1 GGFJ01000312 MBW49453.1 GGFJ01000317 MBW49458.1 GDHF01030870 JAI21444.1 KQ435798 KOX73449.1 GDHF01029457 GDHF01025770 GDHF01020633 GDHF01019694 JAI22857.1 JAI26544.1 JAI31681.1 JAI32620.1 GAMC01015591 JAB90964.1 GDHF01020765 JAI31549.1 GEZM01019409 JAV89647.1 GBXI01014414 JAC99877.1 JRES01000842 KNC27807.1 JXJN01023964 CCAG010009523 CVRI01000010 CRK88716.1 DS235078 EEB11443.1 CRK88717.1 UFQS01000045 UFQT01000045 SSW98418.1 SSX18804.1 GAMC01015588 JAB90967.1 GDHF01029422 GDHF01027328 GDHF01025816 JAI22892.1 JAI24986.1 JAI26498.1 GDHF01025581 JAI26733.1 GEDC01030829 JAS06469.1 GGFM01001332 MBW22083.1 GDHF01033858 JAI18456.1 KB632081 ERL88596.1 GDIP01157117 JAJ66285.1 GDIP01060232 JAM43483.1 GDIP01111275 JAL92439.1 GDIP01125522 JAL78192.1 GDIP01163717 JAJ59685.1 GDIP01171560 JAJ51842.1 GDIP01035935 JAM67780.1 GDIQ01029049 JAN65688.1 GDIP01038384 JAM65331.1 GDIP01160513 JAJ62889.1 GDIP01165912 JAJ57490.1 GDIQ01079777 JAN14960.1 GDIP01165213 JAJ58189.1 GDIP01083233 JAM20482.1 GEDC01028396 JAS08902.1 GDIQ01145714 GDIQ01143911 JAL06012.1 GDIP01155667 JAJ67735.1 GDIP01234762 JAI88639.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000027135
UP000235965
+ More
UP000008237 UP000000311 UP000053097 UP000279307 UP000053825 UP000075809 UP000078540 UP000078542 UP000242457 UP000076502 UP000005203 UP000078492 UP000002358 UP000008820 UP000002320 UP000069940 UP000249989 UP000030765 UP000075884 UP000075880 UP000076408 UP000075886 UP000075885 UP000075881 UP000075883 UP000075920 UP000075902 UP000075840 UP000075882 UP000076407 UP000075903 UP000007062 UP000000673 UP000007266 UP000053105 UP000069272 UP000037069 UP000095300 UP000091820 UP000092445 UP000092460 UP000092443 UP000078200 UP000092444 UP000095301 UP000183832 UP000009046 UP000030742 UP000075900
UP000008237 UP000000311 UP000053097 UP000279307 UP000053825 UP000075809 UP000078540 UP000078542 UP000242457 UP000076502 UP000005203 UP000078492 UP000002358 UP000008820 UP000002320 UP000069940 UP000249989 UP000030765 UP000075884 UP000075880 UP000076408 UP000075886 UP000075885 UP000075881 UP000075883 UP000075920 UP000075902 UP000075840 UP000075882 UP000076407 UP000075903 UP000007062 UP000000673 UP000007266 UP000053105 UP000069272 UP000037069 UP000095300 UP000091820 UP000092445 UP000092460 UP000092443 UP000078200 UP000092444 UP000095301 UP000183832 UP000009046 UP000030742 UP000075900
PRIDE
Interpro
ProteinModelPortal
H9IUM0
A0A212F4H1
A0A194QES9
A0A2A4JTU6
A0A2A4JU20
A0A2H1X5U1
+ More
S4P782 A0A067R8H4 A0A2J7QS75 A0A2J7QS78 A0A310SSF8 E2BUW2 E2AKU8 A0A026WUF4 A0A3L8DEF8 A0A0L7RAQ1 A0A151XJF8 A0A195BFF3 A0A195C2R0 A0A2A3E5I4 A0A154NXX1 A0A088ASY0 A0A151JQB9 K7J2Y9 Q17KE9 B0XJB9 W4VRJ6 A0A182GHW6 A0A084W4P6 A0A182MYW7 A0A182J8B0 A0A182Y3K0 A0A182R0W0 A0A182PCL3 A0A182K3M7 A0A182M4N4 A0A182W6T4 A0A182TF23 A0A182HUD7 A0A2Y9D1F1 A0A182XL18 A0A1S4GYF1 A0A182UPQ9 A0A1Q3F986 A0A1Q3F900 Q7Q461 W5JVX1 D2A126 A0A2M4A3D7 A0A2M4A3M1 A0A2M4A6A5 A0A2M4B8U5 A0A2M4B9F4 A0A0K8U3Y2 A0A0M9A0Q5 A0A0K8U870 W8BMU3 A0A182F152 A0A0K8UY07 A0A1Y1N264 A0A0A1WN99 A0A0L0C6D2 A0A1I8Q9L8 A0A1A9WZI2 A0A1A9ZDA5 A0A1B0C1B0 A0A1A9Y4B4 A0A1A9VDT8 A0A1A9YUH8 A0A1I8N9W6 A0A1J1HMU4 E0VDI7 A0A1J1HL10 A0A336K4B9 W8AY15 A0A0K8U8A5 A0A0K8UJP9 A0A1B6BZM5 A0A2M3Z0N7 A0A0K8TW02 U4UDC5 A0A182R559 A0A0P5DI62 A0A0P5YFK4 A0A0P5URC7 A0A0P5UTS9 A0A0P5E7X3 A0A0P5CNV2 A0A0N8DGI9 A0A0P6INA4 A0A0P5ZVR3 A0A0N8ACM8 A0A0P5CV75 A0A0P6DBC2 A0A0P5CUX4 A0A0P5WPY5 A0A1B6C5X2 A0A0P5NIE0 A0A0P5DXT6 A0A0P4Y4W4
S4P782 A0A067R8H4 A0A2J7QS75 A0A2J7QS78 A0A310SSF8 E2BUW2 E2AKU8 A0A026WUF4 A0A3L8DEF8 A0A0L7RAQ1 A0A151XJF8 A0A195BFF3 A0A195C2R0 A0A2A3E5I4 A0A154NXX1 A0A088ASY0 A0A151JQB9 K7J2Y9 Q17KE9 B0XJB9 W4VRJ6 A0A182GHW6 A0A084W4P6 A0A182MYW7 A0A182J8B0 A0A182Y3K0 A0A182R0W0 A0A182PCL3 A0A182K3M7 A0A182M4N4 A0A182W6T4 A0A182TF23 A0A182HUD7 A0A2Y9D1F1 A0A182XL18 A0A1S4GYF1 A0A182UPQ9 A0A1Q3F986 A0A1Q3F900 Q7Q461 W5JVX1 D2A126 A0A2M4A3D7 A0A2M4A3M1 A0A2M4A6A5 A0A2M4B8U5 A0A2M4B9F4 A0A0K8U3Y2 A0A0M9A0Q5 A0A0K8U870 W8BMU3 A0A182F152 A0A0K8UY07 A0A1Y1N264 A0A0A1WN99 A0A0L0C6D2 A0A1I8Q9L8 A0A1A9WZI2 A0A1A9ZDA5 A0A1B0C1B0 A0A1A9Y4B4 A0A1A9VDT8 A0A1A9YUH8 A0A1I8N9W6 A0A1J1HMU4 E0VDI7 A0A1J1HL10 A0A336K4B9 W8AY15 A0A0K8U8A5 A0A0K8UJP9 A0A1B6BZM5 A0A2M3Z0N7 A0A0K8TW02 U4UDC5 A0A182R559 A0A0P5DI62 A0A0P5YFK4 A0A0P5URC7 A0A0P5UTS9 A0A0P5E7X3 A0A0P5CNV2 A0A0N8DGI9 A0A0P6INA4 A0A0P5ZVR3 A0A0N8ACM8 A0A0P5CV75 A0A0P6DBC2 A0A0P5CUX4 A0A0P5WPY5 A0A1B6C5X2 A0A0P5NIE0 A0A0P5DXT6 A0A0P4Y4W4
Ontologies
PANTHER
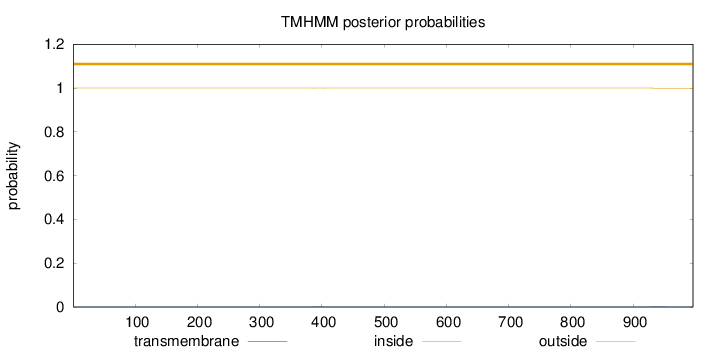
Topology
Length:
996
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01762
Exp number, first 60 AAs:
0.00064
Total prob of N-in:
0.00012
outside
1 - 996
Population Genetic Test Statistics
Pi
222.84313
Theta
210.443813
Tajima's D
0.152898
CLR
0.069664
CSRT
0.405879706014699
Interpretation
Uncertain
